Abstract
The molecular mechanisms governing T helper (Th) cell differentiation and function have revealed a complex network of transcriptional and protein regulators. Cytokines not only initiate the differentiation of CD4 Th cells into subsets but also influence the identity, plasticity and effector function of a T cell. Of the subsets, Th17 cells, named for producing interleukin 17 (IL-17) as their signature cytokine, secrete a cohort of other cytokines, including IL-22, IL-21, IL-10, IL-9, IFNγ, and GM-CSF. In recent years, Th17 cells have emerged as key players in host defense against both extracellular pathogens and fungal infections, but they have also been implicated as one of the main drivers in the pathogenesis of autoimmunity, likely mediated in part by the cytokines that they produce. Advances in high throughput genomic sequencing have revealed unexpected heterogeneity in Th17 cells and, as a consequence, may have tremendous impact on our understanding of their functional diversity. The assortment in gene expression may also identify different functional states of Th17 cells. This review aims to understand the interplay between the cytokine regulators that drive Th17 cell differentiation and functional states in Th17 cells.
Keywords: Th17, T helper cells, inflammation, cytokine signaling
T helper subsets and links to autoimmune inflammation
CD4 T cells are essential architects of host immune defense against pathogens 1, 2. Collectively, their effector function is mediated in part by a compilation of cytokines that directs differentiation, migration, homeostasis, regulation, and inflammation. Initially, CD4 T helper (Th) cells were grouped into two distinct subsets defined by production of unique cytokines: type 1 helper T cells (Th1) produce IFNγ as their signature cytokine and mediate immune responses against intracellular pathogens, and type 2 helper T cells (Th2) secrete interleukin (IL)-4, IL-5 and IL-13 and drive immune responses against extracellular pathogens, like parasites 3. In recent years, the number of unique subsets has grown to include IL-9-producing Th9, follicular T helper cells (Tfh) and IL-17-producing Th17, as well as three subsets of T cells that regulate immune responses, including Type 1 regulatory cells (Tr1), follicular T regulatory cells (TfR) and T regulatory cell (Tregs) ( Figure 1) 4– 6. Each of the effector subsets is not only critical for orchestrating a proper immune response against specific pathogens but is also a major contributor in the pathogenesis of a number of autoimmune inflammatory diseases 7.
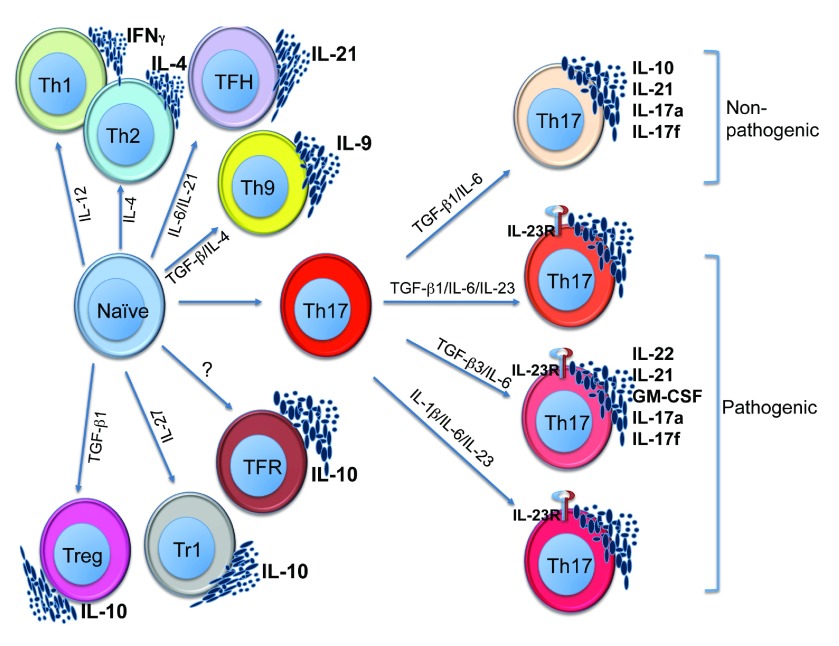
Figure 1. The diversity of CD4 subsets.
CD4 T helper subsets identified with differentiating conditions as well as the signature cytokines they are known to produce. Th17 cells are further subtyped based on cytokine conditions that define pathogenic versus non-pathogenic states.
For a number of years, IL-12-induced Th1 cells were thought to be the main drivers of autoimmunity, based on the findings that IFNγ-secreting CD4 T cells were frequently found at the site of inflammation and treatment with IFNγ led to exacerbated disease in multiple sclerosis patients 8, 9. IL-12 is a heterodimeric cytokine composed of two subunits, IL-12p35 and IL-12p40, and is a critical factor for the differentiation of Th1 cells 10, 11. CD4 T cells express a heterodimeric IL-12 receptor (IL-12R) composed of IL-12Rβ1 and IL-12Rβ2 subunits 12, 13. Upon exposure to IL-12, the master transcription factor Tbx21 is induced, which transactivates IFNγ and the cells differentiate into Th1 cells 14, 15. The importance of Th1 cells in autoimmune diseases was further supported by findings that protection from experimental autoimmune encephalomyelitis (EAE), an animal model of multiple sclerosis, was observed upon neutralization with anti-IL-12p40 or in IL-12p40 –/– mice 16, 17. However, it became clear that Th1 cells may not be the exclusive drivers for autoimmunity when it was discovered that mice lacking critical components of the Th1 differentiation pathway, such as IFNγ, IFNγR, IL-12Rβ2, and IL-12p35, were highly susceptible to EAE, suggesting that Th1 cells may even be protective in autoimmune diseases 18– 22.
Discovery of IL-23- and Th17-associated pathogenic inflammation
In the late 1990s, a novel cytokine called IL-23 that belongs to the IL-12 family of cytokines was discovered 23. Interestingly, similar to the functional IL-12 cytokine, IL-23 had an IL-23 p19 subunit, which combined with the IL-12 p40 subunit of IL-12, to develop a functional heterodimeric cytokine 24. Loss of either IL-23 p19 or IL-12 p40 chains made mice highly resistant to the development of EAE and other autoimmune diseases, suggesting that IL-23 is a cytokine critical for development of autoimmunity 17, 25, 26. However, unlike IL-12, IL-23 did not induce IFNγ production from naïve CD4 T cells 24, 27, but it was suggested that IL-23 may be critical for the generation of IL-17-producing Th17 cells. A series of in vitro studies showed that IL-23 could not induce differentiation of naïve T cells into IL-17-producing Th17 cells. In fact, it was discovered that the receptor for IL-23 was not even expressed on naïve CD4 T cells, suggesting that other cytokines besides IL-23 may be critical for the generation of Th17 cells 28– 30. In fact, we 31 and others 32, 33 showed that Th17 cells are differentiated in the presence of TGF-β1 and IL-6, which resulted in the induction of a unique master transcription factor called RORγt. While IL-23 was not required for the differentiation of Th17 cells, it was revealed to be a critical factor for stabilization of the Th17 phenotype and in evoking pathogenic phenotype in Th17 cells. With ensuing studies it became clear that IL-23, not IL-12, was the critical cytokine for driving autoimmune inflammation. IL-23p19 –/–, IL-12p40 –/– and IL-23R –/– mice 17, 25, 26 were completely protected from developing a number of murine models of autoimmune diseases including EAE, psoriasis, and colitis. Consistently, Genome Wide Association Scans have reported a strong genetic linkage to single nucleotide polymorphisms (SNP) in IL-23 or IL-23R, with increased susceptibility to several human autoimmune diseases 34– 40. However, the clearest role of Th17 cells in human autoimmune diseases was supported by clinical studies where neutralization of IL-17 by an anti-IL-17 antibody (Secukimumab) resulted in clinically beneficial results in a number of human autoimmune diseases, including psoriasis, ankylosing spondylitis, and multiple sclerosis 41– 45.
Heterogeneity within the Th17 subset
Although Th17 cells have become synonymous with autoimmune tissue inflammation, it is now clear that not all Th17 cells are pathogenic or induce tissue inflammation 46. In human inflammatory bowel diseases (IBDs), neutralization of IL-17 or blockade of IL-17 receptor (IL-17Ra) resulted in disease exacerbation, suggesting a possible protective role by Th17 cells 47. IL-17-producing T cells that line the gut mucosa do not induce inflammation but have been shown to be necessary to maintain the barrier function of the gut 48. Commensal bacteria in the gut may play a critical role in the generation of Th17 cells in the lamina propria and, indeed, there is an absence of IL-17-producing cells in the lamina propria of the small intestines in germ-free mice 49, 50. There is also evidence suggesting that IL-17 is required to prevent pathologic gut inflammation in a CD4 T cell-mediated transfer model of colitis, as cells lacking the capacity to produce IL-17, or the lack of IL-17R in recipient mice, resulted in exacerbated colitis 51, 52. These studies alluded to a rather novel concept: that Th17 cells are not uniform in function. In fact, we 53 and others 54, 55 have shown that Th17 cells come in two flavors: one in which they cause pathogenic tissue inflammation and autoimmune disease and the other that is non-pathogenic, in that they fail to provoke autoimmunity, especially in murine T cell models of inflammatory disease ( Figure 1) 53– 55. Th17 cells differentiated in the presence of TGF-β1 and IL-6 56, 57 co-produce IL-17 with IL-10, do not induce tissue inflammation, and in fact may inhibit autoimmune inflammation, and thus are characterized as “non-pathogenic” Th17 cells 55. However, upon exposure to IL-23, a “switch” occurs in the Th17 cell transcriptome, which not only allows for stabilization of the Th17 phenotype but also converts non-pathogenic Th17 cells to become pathogenic 53, 58. These IL-23 experienced Th17 cells have been shown to promote destructive inflammation in numerous T cell-dependent murine models of autoimmunity 53, 58. IL-23 inhibits IL-10 production and instead promotes secretion of IL-22 and GM-CSF, suggesting that IL-23 drives the development of Th17 cells with unique functional properties 59– 61. This raises an important question: how does IL-23 induce pathogenicity in Th17 cells? Our studies revealed that IL-23 mediates important changes in the transcriptome of differentiating Th17 cells 53. Besides the induction of a number of unique transcription factors, IL-23 induces TGF-β3 production in developing Th17 cells 53. We showed that TGF-β3 together with IL-6 in vitro induces differentiation of pathogenic Th17 cells, without any need for further exposure to IL-23 53. Similarly, John O’Shea 54 and Chen Dong’s 62 groups showed that naïve T cells exposed to IL-1β, IL-6 and IL-23 could induce Th17 cells that were highly pathogenic. Thus, by varying the cytokine cocktails in vitro, both pathogenic and non-pathogenic Th17 cells can be generated. Based on these observations, we undertook a systematic transcriptome analysis of Th17 cells in order to develop a novel gene signature that functionally distinguishes Th17 subsets.
Transcriptional gene signatures for pathogenic Th17 cells
When we compared the gene expression profiles of all known possible in vitro differentiation combinations that induce pathogenic and non-pathogenic Th17 cells, we found 434 genes that were differentially expressed between these different Th17 subtypes 53. Of the 434 genes, 233 genes were differentially expressed between highly pathogenic and non-pathogenic Th17 cells 53. Based on the biological function, we identified a representative subset of 23 genes that was highly suggestive of driving pathogenicity 53. Pathogenic Th17 cells induced expression of various effector molecules that have been shown to be pro-inflammatory, such as Cxcl3, Ccl4, Ccl5, Csf2 (GM-CSF) , Il3 (associated with Csf2), Il22, Gzmb (Granzyme B) and, interestingly, transcription factors that are associated with the Th1 phenotype such as Tbx21 (Tbet) and Stat4 53. Conversely, non-pathogenic Th17 cells revealed a gene signature that included molecules associated with regulation, such as Il10 and transcription factors that regulate IL-10 production, such as Ahr and Maf in addition to Ikzf3 (Aiolos) 53. In addition, non-pathogenic Th17 cells express Il1rn (IL-1R antagonist) which might antagonize functions of IL-1 in differentiating Th17 cells into a pathogenic phenotype 53. Based on the comparative gene expression profiles between pathogenic and non-pathogenic Th17 cells, our group identified a gene signature that may confer pathogenic phenotype to Th17 cells 53.
The dichotomous nature of Th17 cells may not be a mere in vitro cytokine artifact but may have occurred naturally as a consequence of evolutionary pressures to defend against different types of pathogens. Federica Sallusto’s group was first to show that human Th17 cells producing IL-10 in conjunction with IL-17 have specificity for Staphylococcus aureus infection 63. Conversely, Th17 cells that do not produce IL-10, but instead produce IFNγ with IL-17, have specificity for Candida albicans infection, suggesting that, evolutionarily, Th17 cells may have diverged to acquire different cytokine profiles, to become more adept in defense against specific pathogens 63. This is in line with clinical observations with immune-deficient patients, where the loss of transcription factor Stat3, which inhibits development of all Th17 cells, results in hyper IgE syndrome and the patients develop rampant C. albicans and S. aureus infections 64. Thus, based on our study, we’ve uncovered an interesting overlap in the gene expression profiles of Th17 cells specific for C. albicans or S. aureus in humans with pathogenic versus non-pathogenic Th17 cells in mice. The gene expression profile revealing an IL-17/IFNγ signature which was specific for C. albicans in humans had similarities to more pathogenic pro-inflammatory murine Th17 cells which cause severe EAE. Conversely, the gene profile for IL-17/IL-10-producing Th17 cells specific for S. aureus were comparable to a more non-pathogenic, regulatory gene signature 53, 54. This was highly suggestive of how evolutionary pressures have fine-tuned different effector cells for clearing different types of pathogens and utilized the same cells for the induction of tissue inflammation or to mediate tissue protection, albeit with small changes in the transcriptome.
Challenges in understanding the functional outcome of Th17 heterogeneity
It has become clear in recent years that Th17 cells may have divergent functions 53. We are just beginning to understand the functional consequences of this extensive heterogeneity of Th17 cells 65. Though gene expression profiling has endowed us with the ability to identify a signature that distinguishes pathogenic from non-pathogenic Th17 cells 53, we do not know how these cells are naturally derived in vivo or what their function is in mediating tissue homeostasis, effector function, inflammation or cancer. For example, do these pathogenic or non-pathogenic Th17 cells develop simultaneously during differentiation in the lymphoid tissue or is there plasticity in the development of Th17 cells such that they can inter-convert based on the environmental cues they receive? Or perhaps there is a sequential development: do non-pathogenic Th17 cells convert into pathogenic Th17 cells during the course of maturation or differentiation? Also, given that the location of the IL-17 producing cells in the peripheral tissue is critical in dictating their function, this raises the issue of how much the peripheral tissue microenvironment alters the developmental programming of Th17 cells. Much remains to be understood in terms of how and why Th17 cells retain heterogeneity and how it influences their functional states.
In recent years, examination of heterogeneity at a single-cell resolution has become possible by high throughput single-cell RNA sequencing of whole genomes and transcriptomes 66, 67. Single-cell RNA sequencing allows for profiling and characterization of expression variability on a genomic scale, which provides us with the ability to correlate this genomic heterogeneity with functional differences in Th17 cells 68, 69. In fact, single-cell RNA sequencing of Th17 cells obtained from different tissues and lymphoid organs is allowing identification of novel regulators of functional states (pathogenic versus non-pathogenic) of Th17 cells (unpublished observation from our lab). Transcriptomic analysis of T cells in the secondary lymphoid organs following activation does provide valuable clues into the differentiation state acquired by the T cells, but it does not identify the functional state that may be attained by Th17 cells upon arrival into the tissue niche. The functional states (pathogenic/non-pathogenic) of the Th17 cells may be partly dependent on the cytokine milieu and tissue microenvironment to which the cells migrate in order to mediate effector functions. Utilizing the pathogenicity gene signature derived from our earlier studies 53 as one of the definable parameters used to analyze single-cell sequence data, our lab has discovered that the functional states of Th17 cells may be in constant flux as the T cells mediate tissue inflammation (unpublished observation). Uncovering key regulators that control effector functions of Th17 cells may permit novel treatment approaches for therapeutically inhibiting inflammation without affecting the protective functions of Th17 cells.
However, assigning function to these novel regulators will require genetic manipulation undertaken at a large scale. Unfortunately, the only way to confirm the function of a gene is through the use of knockout mice or genetic knockdowns in the cells and disease models 65, 70. The use of viral vectors or transfection-based si-RNA delivery was not effective in this endeavor, due to the changes in either the differentiation or cell viability induced by these manipulations 71, 72. Also, to generate a knockout mouse of every novel regulator identified at a single-cell level is an impossible undertaking. To bypass these obvious limitations, our lab in collaboration with Hongkun Park’s lab has developed a novel system of silicon nanowire perturbations where newly discovered candidate genes can be knocked-down at a large scale, which has improved the process of functional validation 65. Silicon nanowire perturbation allows for the delivery of siRNA effectively and efficiently into native T cells without the burden of activation or differentiation 65, 73, 74.
The future
Armed with next-generation sequencing and silicon nanowire knockdowns, the pathogenic potential of subpopulations within Th17 cells can be revealed and novel regulators that may drive functional heterogeneity can be effectively established. Understanding the epigenetic and transcriptional controls of various functional states of Th17 cells will undoubtedly reveal new treatment paradigms for autoimmune diseases as well as give us deeper insight into the complex network that drives inflammatory versus tissue-protective functions of Th17 cells.
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
v1; ref status: indexed
References
- 1. Khader SA, Gopal R: IL-17 in protective immunity to intracellular pathogens. Virulence. 2010;1(5):423–427. 10.4161/viru.1.5.12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khader SA, Guglani L, Rangel-Moreno J, et al. : IL-23 is required for long-term control of Mycobacterium tuberculosis and B cell follicle formation in the infected lung. J Immunol. 2011;187(10):5402–5407. 10.4049/jimmunol.1101377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mosmann TR, Coffman RL: TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. 10.1146/annurev.iy.07.040189.001045 [DOI] [PubMed] [Google Scholar]
- 4. King C, Tangye SG, Mackay CR: T follicular helper (T FH) cells in normal and dysregulated immune responses. Annu Rev Immunol. 2008;26:741–766. 10.1146/annurev.immunol.26.021607.090344 [DOI] [PubMed] [Google Scholar]
- 5. Zhu J, Paul WE: Heterogeneity and plasticity of T helper cells. Cell Res. 2010;20(1):4–12. 10.1038/cr.2009.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu J, Yamane H, Paul WE: Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. 2010;28:445–489. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cox CA, Shi G, Yin H, et al. : Both Th1 and Th17 are immunopathogenic but differ in other key biological activities. J Immunol. 2008;180(11):7141–22. 10.4049/jimmunol.180.11.7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zamvil SS, Steinman L: The T lymphocyte in experimental allergic encephalomyelitis. Annu Rev Immunol. 1990;8:579–621. 10.1146/annurev.iy.08.040190.003051 [DOI] [PubMed] [Google Scholar]
- 9. Panitch HS, Hirsch RL, Haley AS, et al. : Exacerbations of multiple sclerosis in patients treated with gamma interferon. Lancet. 1987;1(8538):893–895. 10.1016/S0140-6736(87)92863-7 [DOI] [PubMed] [Google Scholar]
- 10. Wolf SF, Temple PA, Kobayashi M, et al. : Cloning of cDNA for natural killer cell stimulatory factor, a heterodimeric cytokine with multiple biologic effects on T and natural killer cells. J Immunol. 1991;146(9):3074–3081. [PubMed] [Google Scholar]
- 11. Stern AS, Magram J, Presky DH: Interleukin-12 an integral cytokine in the immune response. Life Sci. 1996;58(8):639–654. 10.1016/S0024-3205(96)80003-8 [DOI] [PubMed] [Google Scholar]
- 12. Chua AO, Chizzonite R, Desai BB, et al. : Expression cloning of a human IL-12 receptor component. A new member of the cytokine receptor superfamily with strong homology to gp130. J Immunol. 1994;153(1):128–136. [PubMed] [Google Scholar]
- 13. Presky DH, Yang H, Minetti LJ, et al. : A functional interleukin 12 receptor complex is composed of two beta-type cytokine receptor subunits. Proc Natl Acad Sci U S A. 1996;26(24):14002–7. 10.1073/pnas.93.24.14002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gately MK, Renzetti LM, Magram J, et al. : The interleukin-12/interleukin-12-receptor system: role in normal and pathologic immune responses. Annu Rev Immunol. 1998;16:495–521. 10.1146/annurev.immunol.16.1.495 [DOI] [PubMed] [Google Scholar]
- 15. Szabo SJ, Kim ST, Costa GL, et al. : A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–669. 10.1016/S0092-8674(00)80702-3 [DOI] [PubMed] [Google Scholar]
- 16. Brok Herbert PM, van Meurs M, Blezer E, et al. : Prevention of experimental autoimmune encephalomyelitis in common marmosets using an anti-IL-12p40 monoclonal antibody. J Immunol. 2002;169(11):6554–6563. 10.4049/jimmunol.169.11.6554 [DOI] [PubMed] [Google Scholar]
- 17. Langrish CL, Chen Y, Blumenschein WM, et al. : IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201(2):233–240. 10.1084/jem.20041257 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Gran B, Zhang G, Yu S, et al. : IL-12p35-deficient mice are susceptible to experimental autoimmune encephalomyelitis: evidence for redundancy in the IL-12 system in the induction of central nervous system autoimmune demyelination. J Immunol. 2002;169(12):7104–7110. 10.4049/jimmunol.169.12.7104 [DOI] [PubMed] [Google Scholar]
- 19. Becher B, Durell BG, Noelle RJ: Experimental autoimmune encephalitis and inflammation in the absence of interleukin-12. J Clin Invest. 2002;110(4):493–497. 10.1172/JCI15751 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 20. Zhang G, Gran B, Yu S, et al. : Induction of experimental autoimmune encephalomyelitis in IL-12 receptor-beta 2-deficient mice: IL-12 responsiveness is not required in the pathogenesis of inflammatory demyelination in the central nervous system. J Immunol. 2003;170(4):2153–2160. 10.4049/jimmunol.170.4.2153 [DOI] [PubMed] [Google Scholar]
- 21. Willenborg DO, Fordham S, Bernard CC, et al. : IFN-gamma plays a critical down-regulatory role in the induction and effector phase of myelin oligodendrocyte glycoprotein-induced autoimmune encephalomyelitis. J Immunol. 1996;157(8):3223–3227. [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Ferber IA, Brocke S, Taylor-Edwards C, et al. : Mice with a disrupted IFN-gamma gene are susceptible to the induction of experimental autoimmune encephalomyelitis (EAE). J Immunol. 1996;156(1):5–7. [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Oppmann B, Lesley R, Blom B, et al. : Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13(5):715–725. 10.1016/S1074-7613(00)00070-4 [DOI] [PubMed] [Google Scholar]
- 24. Langrish CL, McKenzie BS, Wilson NJ, et al. : IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol Rev. 2004;202(1):96–105. 10.1111/j.0105-2896.2004.00214.x [DOI] [PubMed] [Google Scholar]
- 25. Cua DJ, Sherlock J, Chen Y, et al. : Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421(6924):744–748. 10.1038/nature01355 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Croxford AL, Mair F, Becher B: IL-23: one cytokine in control of autoimmunity. Eur J Immunol. 2012;42(9):2263–2273. 10.1002/eji.201242598 [DOI] [PubMed] [Google Scholar]
- 27. Aggarwal S, Ghilardi N, Xie M, et al. : Interleukin-23 promotes a distinct CD 4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278(3):1910–1914. 10.1074/jbc.M207577200 [DOI] [PubMed] [Google Scholar]
- 28. Diveu C, McGeachy MJ, Cua DJ: Cytokines that regulate autoimmunity. Curr Opin Immunol. 2008;20(6):663–668. 10.1016/j.coi.2008.09.003 [DOI] [PubMed] [Google Scholar]
- 29. Murphy CA, Langrish CL, Chen Y, et al. : Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198(12):1951–7. 10.1084/jem.20030896 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Uhlig HH, McKenzie BS, Hue S, et al. : Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25(2):309–318. 10.1016/j.immuni.2006.05.017 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Bettelli E, Carrier Y, Gao W, et al. : Reciprocal developmental pathways for the generation of pathogenic effector T H17 and regulatory T cells. Nature. 2006;441(7090):235–238. 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Mangan PR, Harrington LE, O’Quinn DB, et al. : Transforming growth factor-beta induces development of the T H17 lineage. Nature. 2006;441(7090):231–234. 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 33. Veldhoen M, Hocking RJ, Atkins CJ, et al. : TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24(2):179–189. 10.1016/j.immuni.2006.01.001 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Cargill M, Schrodi SJ, Chang M, et al. : A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80(2):273–90. 10.1086/511051 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Rahman P, Inman RD, Gladman DD, et al. : Association of interleukin-23 receptor variants with ankylosing spondylitis. Arthritis Rheum. 2008;58(4):1020–1025. 10.1002/art.23389 [DOI] [PubMed] [Google Scholar]
- 36. Rahman P, Inman RD, Maksymowych WP, et al. : Association of interleukin 23 receptor variants with psoriatic arthritis. J Rheumatol. 2009;36(1):137–140. 10.3899/jrheum.080458 [DOI] [PubMed] [Google Scholar]
- 37. Huber AK, Jacobson EM, Jazdzewski K, et al. : Interleukin (IL)-23 receptor is a major susceptibility gene for Graves' ophthalmopathy: the IL-23/T-helper 17 axis extends to thyroid autoimmunity. J Clin Endocrinol Metab. 2008;93(3):1077–1081. 10.1210/jc.2007-2190 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Ali S, Srivastava AK, Chopra R, et al. : IL12B SNPs and copy number variation in IL23R gene associated with susceptibility to leprosy. J Med Genet. 2013;50(1):34–42. 10.1136/jmedgenet-2012-101214 [DOI] [PubMed] [Google Scholar]
- 39. Núñez C, Dema B, Cénit MC, et al. : IL23R: a susceptibility locus for celiac disease and multiple sclerosis? Genes Immun. 2008;9(4):289–293. 10.1038/gene.2008.16 [DOI] [PubMed] [Google Scholar]
- 40. Eirís N, González-Lara L, Santos-Juanes J, et al. : Genetic variation at IL12B, IL23R and IL23A is associated with psoriasis severity, psoriatic arthritis and type 2 diabetes mellitus. J Dermatol Sci. 2014;75(3):167–172. 10.1016/j.jdermsci.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 41. Hofstetter HH, Ibrahim SM, Koczan D, et al. : Therapeutic efficacy of IL-17 neutralization in murine experimental autoimmune encephalomyelitis. Cell Immunol. 2005;237(2):123–130. 10.1016/j.cellimm.2005.11.002 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Luger D, Silver PB, Tang J, et al. : Either a Th17 or a Th1 effector response can drive autoimmunity: conditions of disease induction affect dominant effector category. J Exp Med. 2008;205(4):799–810. 10.1084/jem.20071258 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Genovese MC, Van den Bosch F, Roberson SA, et al. : LY2439821, a humanized anti-interleukin-17 monoclonal antibody, in the treatment of patients with rheumatoid arthritis: A phase I randomized, double-blind, placebo-controlled, proof-of-concept study. Arthritis Rheum. 2010;62(4):929–939. 10.1002/art.27334 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Huh JR, Leung MW, Huang P, et al. : Digoxin and its derivatives suppress T H17 cell differentiation by antagonizing RORγt activity. Nature. 2011;472(7344):486–490. 10.1038/nature09978 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Xiao S, Yosef N, Yang J, et al. : Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40(4):477–489. 10.1016/j.immuni.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maddur MS, Miossec P, Kaveri SV, et al. : Th17 cells: biology, pathogenesis of autoimmune and inflammatory diseases, and therapeutic strategies. Am J Pathol. 2012;181(1):8–18. 10.1016/j.ajpath.2012.03.044 [DOI] [PubMed] [Google Scholar]
- 47. Hueber W, Sands BE, Lewitzky S, et al. : Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61(12):1693–1700. 10.1136/gutjnl-2011-301668 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Kamada N, Seo S, Chen GY, et al. : Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- 49. Atarashi K, Nishimura J, Shima T, et al. : ATP drives lamina propria T H17 cell differentiation. Nature. 2008;455(7214):808–812. 10.1038/nature07240 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Ivanov II, Frutos Rde L, Manel N, et al. : Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 51. O’Connor W, Jr, Kamanaka M, Booth CJ, et al. : A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol. 2009;10(6):603–609. 10.1038/ni.1736 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. O’Connor W, Jr, Zenewicz LA, Flavell RA: The dual nature of T H17 cells: shifting the focus to function. Nat Immunol. 2010;11(6):471–476. 10.1038/ni.1882 [DOI] [PubMed] [Google Scholar]
- 53. Lee Y, Awasthi A, Yosef N, et al. : Induction and molecular signature of pathogenic T H17 cells. Nat Immunol. 2012;13(10):991–999. 10.1038/ni.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Ghoreschi K, Laurence A, Yang X, et al. : Generation of pathogenic T H17 cells in the absence of TGF-β signalling. Nature. 2010;467(7318):967–971. 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. McGeachy MJ, Bak-Jensen KS, Chen Y, et al. : TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T H-17 cell-mediated pathology. Nat Immunol. 2007;8(12):1390–1397. 10.1038/ni1539 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. McGeachy MJ, Cua DJ: Th17 cell differentiation: the long and winding road. Immunity. 2008;28(4):445–453. 10.1016/j.immuni.2008.03.001 [DOI] [PubMed] [Google Scholar]
- 57. Korn T, Bettelli E, Oukka M, et al. : IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- 58. Jäger A, Dardalhon V, Sobel RA, et al. : Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J Immunol. 2009;183(11):7169–7177. 10.4049/jimmunol.0901906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Codarri L, Gyülvészi G, Tosevski V, et al. : RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12(6):560–567. 10.1038/ni.2027 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Duvallet E, Semerano L, Assier E, et al. : Interleukin-23: a key cytokine in inflammatory diseases. Ann Med. 2011;43(7):503–511. 10.3109/07853890.2011.577093 [DOI] [PubMed] [Google Scholar]
- 61. El-Behi M, Ciric B, Dai H, et al. : The encephalitogenicity of T H17 cells is dependent on IL-1-and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12(6):568–575. 10.1038/ni.2031 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 62. Chung Y, Chang SH, Martinez GJ, et al. : Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30(4):576–587. 10.1016/j.immuni.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Zielinski CE, Mele F, Aschenbrenner D, et al. : Pathogen-induced human T H17 cells produce IFN-γ or IL-10 and are regulated by IL-1β. Nature. 2012;484(7395):514–518. 10.1038/nature10957 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Ma CS, Chew GY, Simpson N, et al. : Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205(7):1551–1557. 10.1084/jem.20080218 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Yosef N, Shalek AK, Gaublomme JT, et al. : Dynamic regulatory network controlling T H17 cell differentiation. Nature. 2013;496(7446):461–468. 10.1038/nature11981 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Kalisky T, Blainey P, Quake SR: Genomic analysis at the single-cell level. Annu Rev Genet. 2011;45:431–445. 10.1146/annurev-genet-102209-163607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Blainey PC, Quake SR: Dissecting genomic diversity, one cell at a time. Nat Methods. 2014;11(1):19–21. 10.1038/nmeth.2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Trombetta JJ, Gennert D, Lu D, et al. : Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr Protoc Mol Biol. 2014;107:4.22.1–4.22.17. 10.1002/0471142727.mb0422s107 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Shalek AK, Satija R, Adiconis X, et al. : Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–240. 10.1038/nature12172 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 70. Yosef N, Zalckvar E, Rubinstein AD, et al. : ANAT: a tool for constructing and analyzing functional protein networks. Sci Signal. 2011;4(196):pl1. 10.1126/scisignal.2001935 [DOI] [PubMed] [Google Scholar]
- 71. Dardalhon V, Herpers B, Noraz N, et al. : Lentivirus-mediated gene transfer in primary T cells is enhanced by a central DNA flap. Gene Ther. 2001;8(3):190–198. 10.1038/sj.gt.3301378 [DOI] [PubMed] [Google Scholar]
- 72. McManus MT, Haines BB, Dillon CP, et al. : Small interfering RNA-mediated gene silencing in T lymphocytes. J Immunol. 2002;169(10):5754–5760. 10.4049/jimmunol.169.10.5754 [DOI] [PubMed] [Google Scholar]
- 73. Shalek AK, Robinson JT, Karp ES, et al. : Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc Natl Acad Sci U S A. 2010;107(5):1870–1875. 10.1073/pnas.0909350107 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Shalek AK, Gaublomme JT, Wang L, et al. : Nanowire-mediated delivery enables functional interrogation of primary immune cells: application to the analysis of chronic lymphocytic leukemia. Nano lett. 2012;12(12):6498–6504. 10.1021/nl3042917 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation