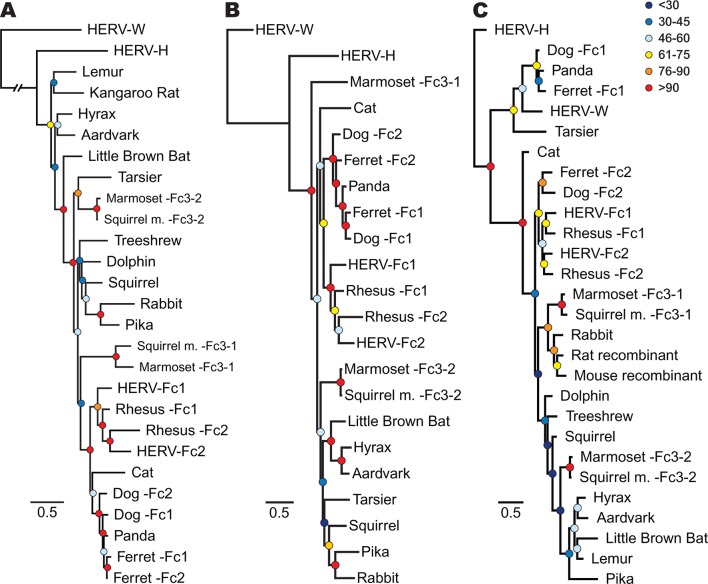
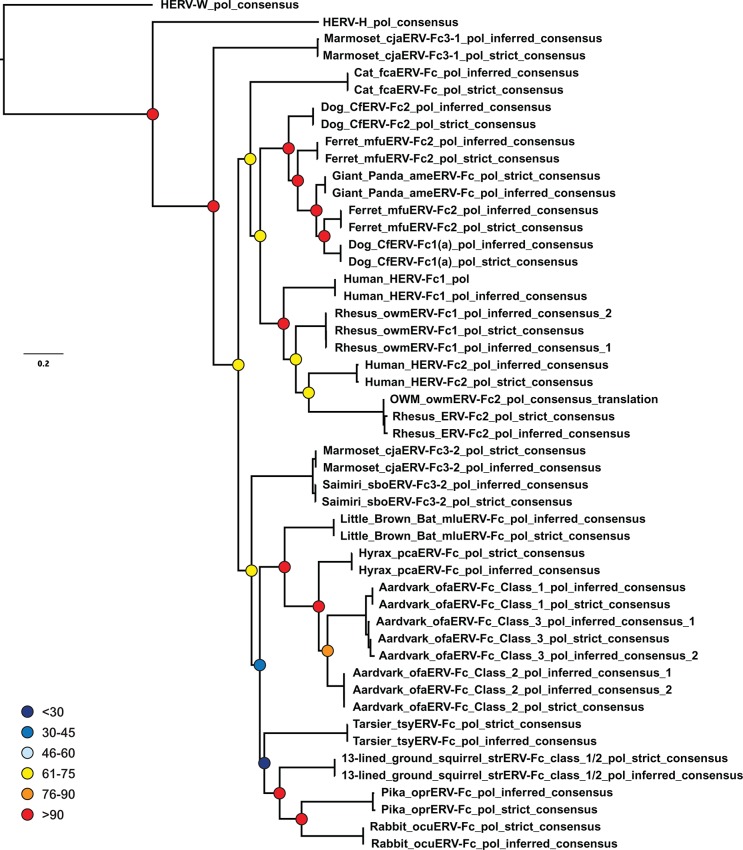
Figure 4. Phylogenetic relationship between ERV-Fc sequences.
Maximum likelihood amino acid trees of (A) Gag (B) Pol and (C) TM generated using the LG substitution matrix. In each panel, HERV-H and HERV-W sequences were included as outgroups. Boostrap confidence values of nodes are depicted by colored spheres. In order to save space, a distance of approximately 0.6 was removed from the HERV-W outgroup branch in the Gag phylogeny (A), as indicated by the broken line.
DOI: http://dx.doi.org/10.7554/eLife.12704.011