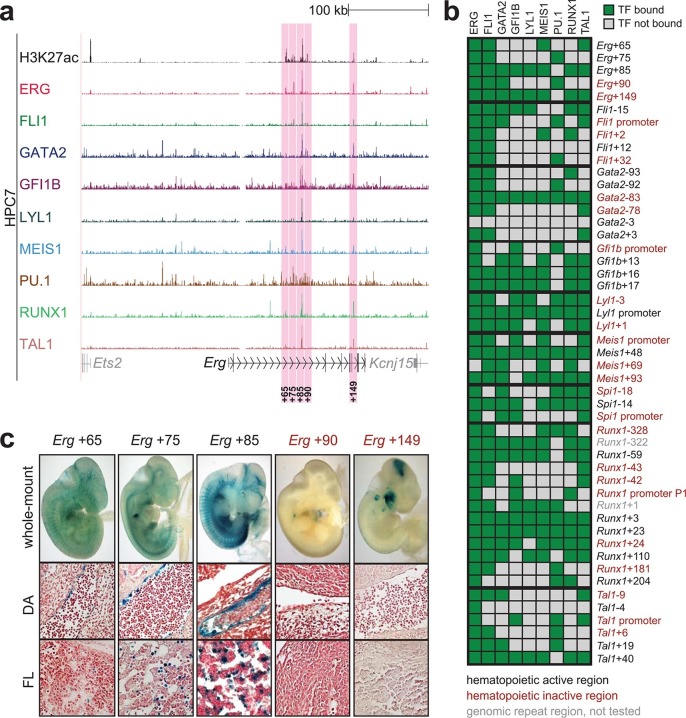
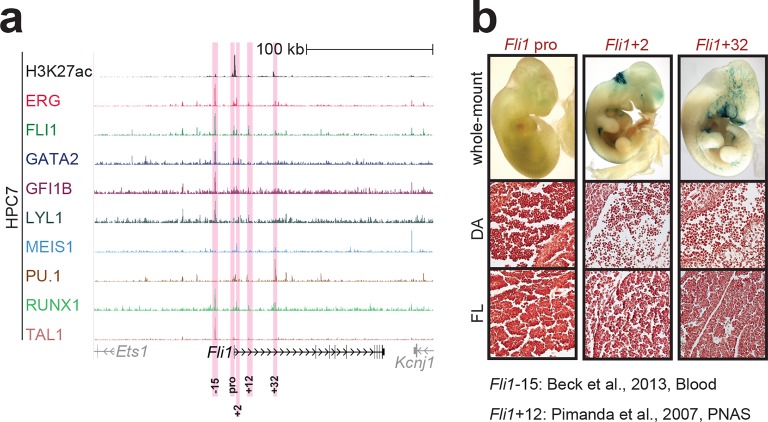
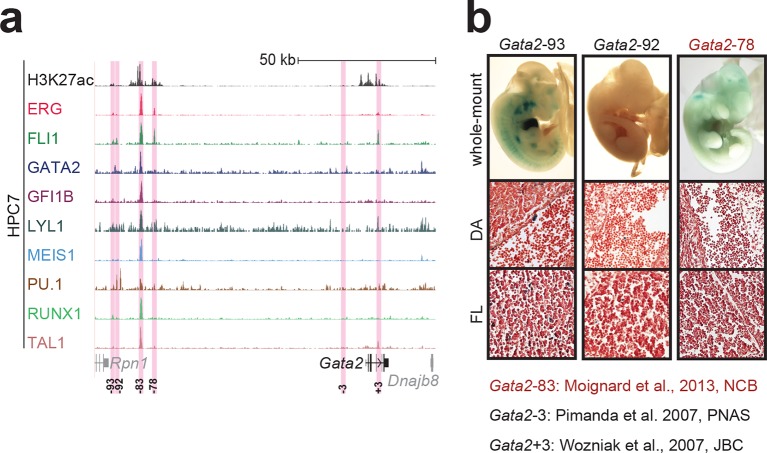
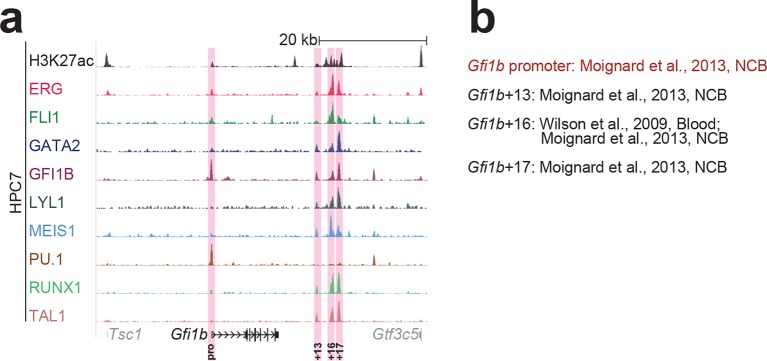
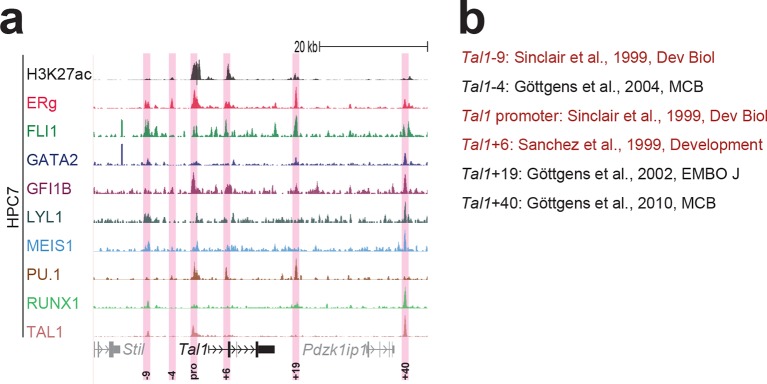
Figure 1. Identification of haematopoietic active cis-regulatory regions.
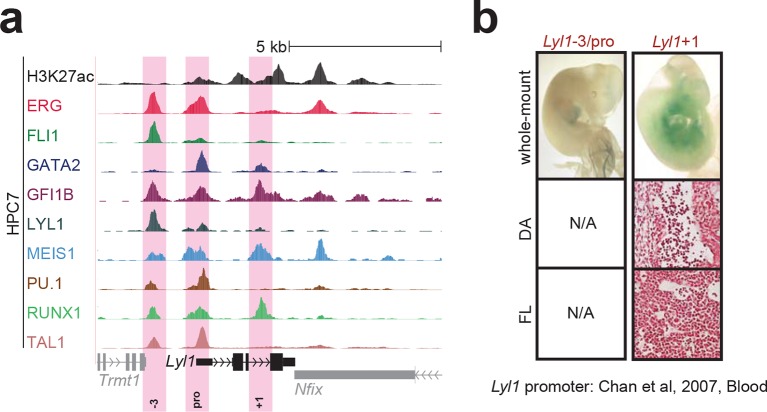
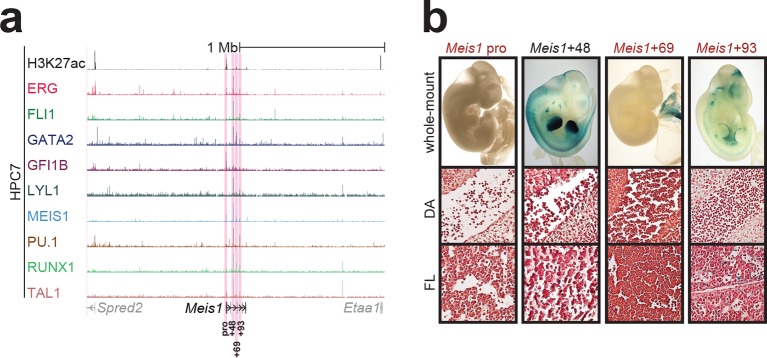
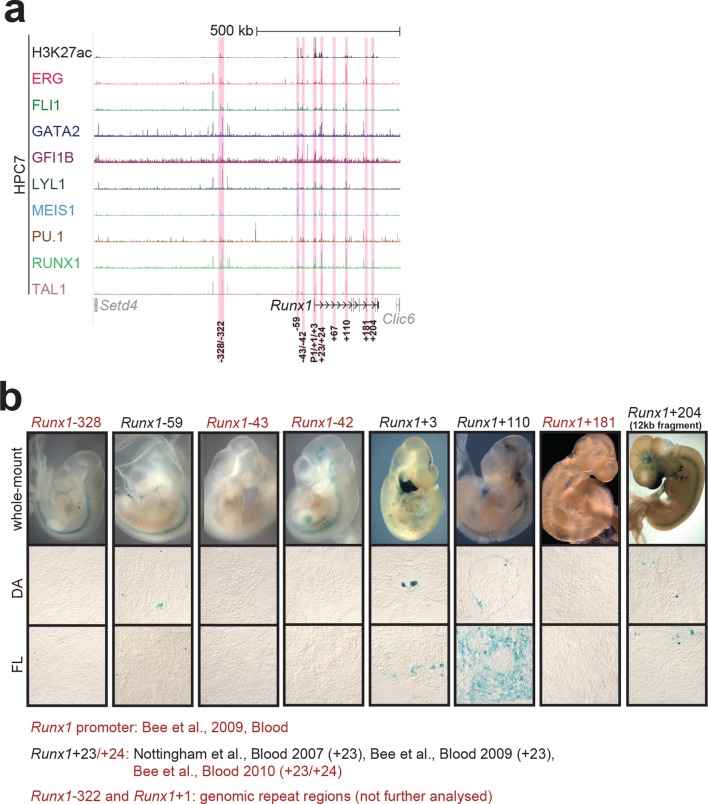
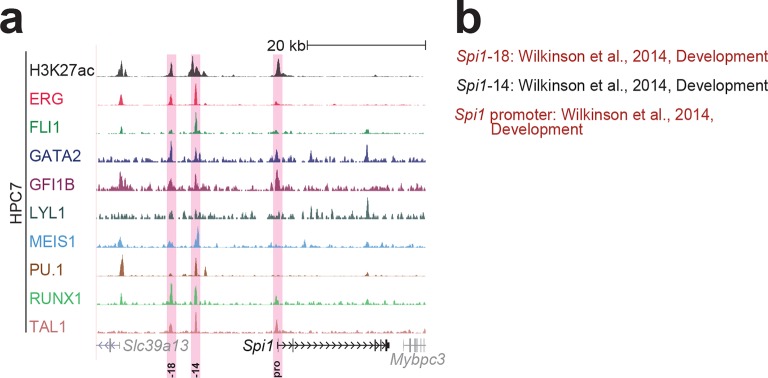
(a) UCSC screenshot of the Erg gene locus for ChIP-Sequencing data for nine haematopoietic TFs (ERG, FLI1, GATA2, GFI1B, LYL1, MEIS1, PU.1, RUNX1 and TAL1 [Wilson et al., 2010]) and for H3K27ac (Calero-Nieto et al., 2014) in HPC7 cells. Highlighted are all regions of the Erg gene locus that are acetylated at H3K27 and are bound by three or more TFs. Numbers indicate the distance (in kb) from the ATG start codon. (b) Summary of the identification of candidate cis-regulatory regions for all nine TFs and subsequent analysis in transgenic mouse assays. The inspection of the nine gene loci and the application of the selection criteria (≥3 TFs bound and H3K27ac) identified a total of 49 candidate cis-regulatory regions. The heatmap shows the binding pattern of the nine TFs to all candidate regulatory elements in HPC7 cells: green = bound, grey = unbound. Haematopoietic activity in E11.5 transgenic mice is indicated by the font color: black = active, red = not active. Grey indicates genomic repeat regions that were not tested in transgenic mice. Detailed experimental data corresponding to the summary heatmap can be found in Figure 1 and Figure 1—figure supplement 1–8. (c) Haematopoietic activity of the five candidate Erg cis-regulatory regions was determined in E11.5 transgenic mouse assays. Shown are X-Gal-stained whole-mount embryos and paraffin sections of the dorsal aorta (DA, ventral side on the left/top) and foetal liver (FL), sites of definitive haematopoiesis. Colour coding as in B.
DOI: http://dx.doi.org/10.7554/eLife.11469.003