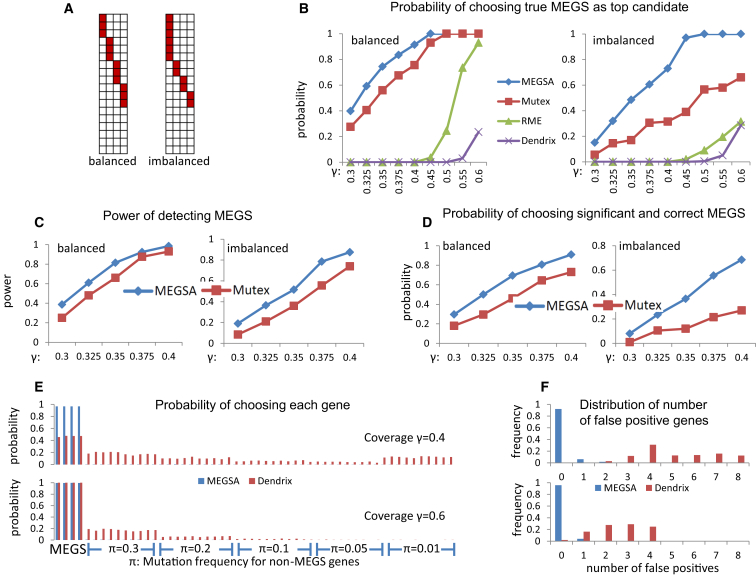
Figure 3.
Performance Comparison of Methods for Detecting Mutually Exclusive Gene Sets on Simulated Datasets
In all simulations, we have 54 genes with 50 being randomly simulated with specific mutation frequencies and 4 genes as MEGS.
(A) Balanced and imbalanced MEGS with four genes. In imbalanced MEGS, the mutational frequencies have a ratio 3:1:1:1.
(B) Probability of ranking the exact MEGS as the top candidate. The X-coordinate is the coverage (γ) of simulated MEGS.
(C) Power of detecting MEGS via MEGSA and Mutex.
(D) Probability that the identified top MEGS is statistically significant and identical to the true MEGS. Coverage (γ) of MEGS ranges from 0.3 to 0.4.
(E) Probability of choosing each gene in the identified top MEGS by MEGSA and Dendrix. The top figure is based on a MEGS with coverage γ = 0.4 and the bottom figure based on coverage γ = 0.6. π is the mutation frequencies for the 50 non-MEGS genes. The first four are MEGS genes and the rest are non-MEGS genes.
(F) The distribution of the number of falsely detected genes for the top MEGS identified by in MEGSA and Dendrix. MEGSA had few false-positive genes whereas Dendrix detected many false-positive genes.