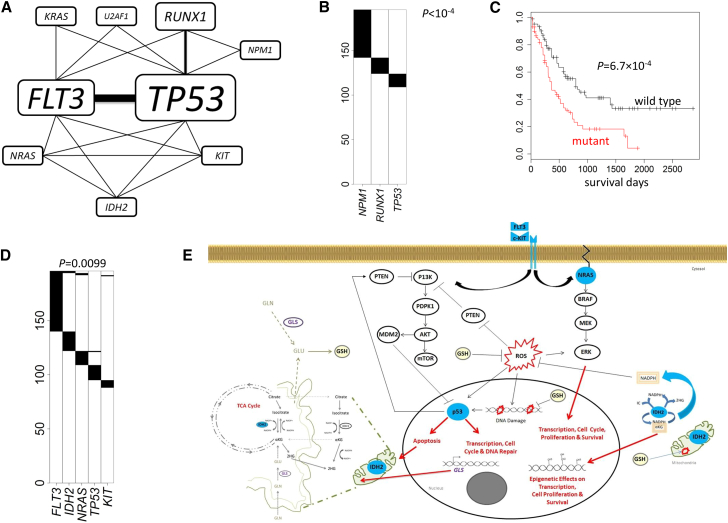
Figure 5.
Analysis Results for TCGA Acute Myeloid Leukemia Whole-Exome Sequencing Data
p values were adjusted for multiple testing for all reported MEGS.
(A) A network constructed based on the five significant MEGS. Thickness of the edges and sizes of the gene labels are proportional to the times in the detected MEGS.
(B) The most significant MEGS with three genes: NPM1, RUNX1, and TP53.
(C) The mutation status of the triplet (NPM1, RUNX1, and TP53) is strongly associated with survival.
(D) A significant MEGS with five genes.
(E) Illustration showing MEGS pattern including protein products (colored blue) of FLT3, IDH2, KIT, NRAS, and TP53 in their relevant biological pathways. Connections including activation (lines with arrow) and inhibition (bar-headed lines) as well as end biological effects between the gene products are indicated. IDH2, which locates to the mitochondria, is shown outside (or in-part) of the illustrated organelle for clarity and only the relevant components of glutamine (GLN) and glutathione (GSH) metabolism and TCA cycle are indicated. PI3K pathway (receptor tyrosine kinases [RTK], FLIT3, and KIT), MAPK/ERK pathway (NRAS). Abbreviations are as follows: ROS, reactive oxygen species, αKG, alpha-ketoglutarate; 2HG, 2-hydroxyglutarate; TCA, tricarboxylic acid cycle; GLN, glutamine; GLU, glutamate; and GLS, glutaminase 2; GSH (glutathione).