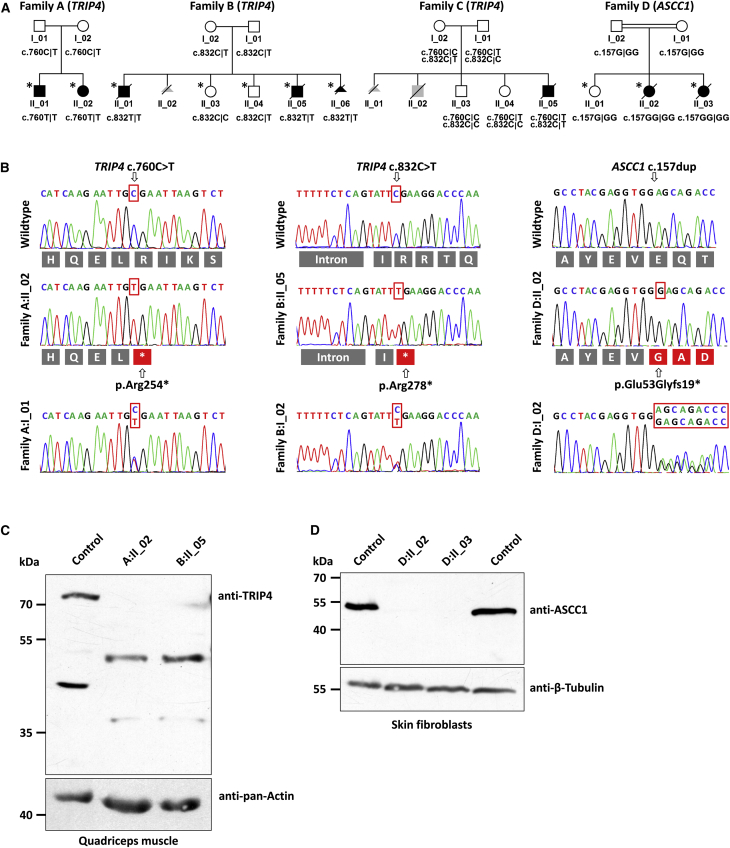
Figure 2.
Pedigrees of the Families and Molecular Genetic Findings
(A) The pedigrees of all investigated families with their respective genotypes are depicted below the symbols. Individuals marked with an asterisk were used for autozygosity mapping.
(B) Variants identified by WES were verified by Sanger sequencing for segregation in all family members. Below the electropherograms, the reading frame of the respective amino acids is provided in the three letter code. Both TRIP4 mutations resulted in a premature termination codon, whereas the ASCC1 mutation led to a frameshift with a termination codon after insertion of 19 non-original amino acids.
(C) Western blot of a muscle protein extract from two individuals with a TRIP4 mutation and from a control individual. The premature termination codons in both affected children lead to upregulation of an alternative splice isoform at ∼53 kDa that excludes both mutant positions. Anti-pan-Actin band density was used as a loading control.
(D) Western blot of a protein extract from cultured fibroblasts of both children from family D and of two control individuals. The blot demonstrates the complete absence of the ASCC1 band in the affected children. β-tubulin band density was used as a loading control.