Abstract
Therapy resistance remains a major problem in estrogen receptor-α (ERα)-positive breast cancer. A subgroup of ERα-positive breast cancer is characterized by mosaic presence of a minor population of ERα-negative cancer cells expressing the basal cytokeratin-5 (CK5). These CK5-positive cells are therapy resistant and have increased tumor-initiating potential. Although a series of reports document induction of the CK5-positive cells by progestins, it is unknown if other 3-ketosteroids share this ability. We now report that glucocorticoids and mineralocorticoids effectively expand the CK5-positive cell population. CK5-positive cells induced by 3-ketosteroids lacked ERα and progesterone receptors, expressed stem cell marker, CD44, and displayed increased clonogenicity in soft agar and broad drug-resistance in vitro and in vivo. Upregulation of CK5-positive cells by 3-ketosteroids required induction of the transcriptional repressor BCL6 based on suppression of BCL6 by two independent BCL6 small hairpin RNAs or by prolactin. Prolactin also suppressed 3-ketosteroid induction of CK5+ cells in T47D xenografts in vivo. Survival analysis with recursive partitioning in node-negative ERα-positive breast cancer using quantitative CK5 and BCL6 mRNA or protein expression data identified patients at high or low risk for tumor recurrence in two independent patient cohorts. The data provide a mechanism by which common pathophysiological or pharmacologic elevations in glucocorticoids or other 3-ketosteroids may adversely affect patients with mixed ERα+/CK5+ breast cancer. The observations further suggest a cooperative diagnostic utility of CK5 and BCL6 expression levels and justify exploring efficacy of inhibitors of BCL6 and 3-ketosteroid receptors for a subset of ERα-positive breast cancers.
Introduction
Estrogen receptor-α positive (ERα+) disease comprises 70–80% of all newly diagnosed invasive breast carcinomas.1, 2 Despite widespread use of adjuvant anti-estrogen and chemotherapy, approximately one in four patients with ERα+ breast cancer will experience recurrence of therapy-resistant disease.3, 4 Although selection for cancer cells with genomic or epigenetic alterations may underlie a portion of subsequent therapy-resistant disease, recurrence of some ERα+ breast cancers may alternatively result from the expansion of a pre-existing cell population that is intrinsically resistant to adjuvant therapy.5 Tumor marker-based classification of ERα+ breast cancer into therapy-relevant subtypes remains an ongoing effort. Protein expression phenotyping of breast cancer includes hormone receptor status (ERα, progesterone receptor (PR)), Her2 status, proliferation status (Ki67) and basal cytokeratin-5 (CK5) expression.6 Luminal A breast cancer (ERα+/CK5-/Her2-/Ki67low) is associated with favorable outcome with adjuvant anti-estrogen therapy. Luminal B breast cancer (ERα+/CK5-/Her2±/Ki67high) confers poor prognosis but patients do benefit from anti-estrogen therapy. An emerging subgroup of ERα+ breast cancer display predominantly ERα+/CK5− cells and mosaic presence of a subpopulation of ERα−/CK5+ cells, referred to as ‘mixed luminal and basal'7 or ‘Luminobasal' breast cancer.8, 9 These tumors are thought to develop resistance to common adjuvant therapies through expansion of the ERα−/CK5+ cell population.8, 9, 10
CK5 is expressed in baso-luminal precursor cells of healthy breast epithelia and is a commonly used marker for tumors within the basal-like subtype of triple-negative breast cancer (TNBC) but has generally been considered absent in ERα+ breast cancers.11, 12 However, 10–50% of ERα+ breast cancers contain a rare population of CK5+ cells.7, 8, 9, 13 This CK5+ cell population is further enriched in ERα+ tumors of patients who have experienced recurrence following chemo or anti-estrogen therapies.8, 13 In addition, in vitro and in vivo data indicate that the CK5+ cells display reduced sensitivity to adjuvant therapy and increased tumor-initiating potential,8, 13 further supporting a role of CK5+ cells in the recurrence of ERα+ breast cancer. A series of reports have established that PR activation expands the CK5+ cell population in experimental models of ERα+ breast cancer.14, 15, 16, 17, 18 It remains unknown, however, whether related members of the 3-ketosteroid nuclear receptor family, including receptors for glucocorticoids, mineralocorticoids and androgens, also stimulate expansion of this therapy-resistant CK5+ cell population. In fact, limited evidence has indicated that induction of CK5+ cells in ERα+ breast cancer is a unique effect of progestin (Prg) that is not mimicked by other steroids, including glucocorticoids.19 The four members of the 3-ketosteroid receptor family bind to overlapping but distinct DNA response elements, are variably expressed in cell populations, and interact differentially with co-regulators, thus exerting both similar and distinct biological effects in breast cancer cells.20, 21, 22, 23 Due to the wide use of glucocorticoids for a range of common conditions that frequently are comorbidities in patients with breast cancer, their administration at high doses with adjuvant chemotherapy to attenuate nausea and off-target toxicity, as well as their frequent use in metastatic cases for symptom management, we undertook a systematic analysis to determine whether 3-ketosteroids other than progesterone could expand the CK5+ cell population in ERα+ breast cancer.
The present study provides novel evidence that in ERα+ breast cancer several 3-ketosteroids, including glucocorticoids and mineralocorticoids, mimic Prg-induced expansion of therapy-resistant CK5+ cells through a mechanism that requires the transcriptional repressor and oncogene BCL6. The data are supported by experimental studies of ERα+ breast cancer cell lines in vitro and in vivo. Further clinical significance is provided by the strong association between early disease recurrence and combined CK5 and BCL6 mRNA or protein expression levels in patients with ERα+ breast cancer.
Results
Glucocorticoid and aldosterone mimic Prg induction of a CK5+ cell population in ERα+ breast cancer cell lines
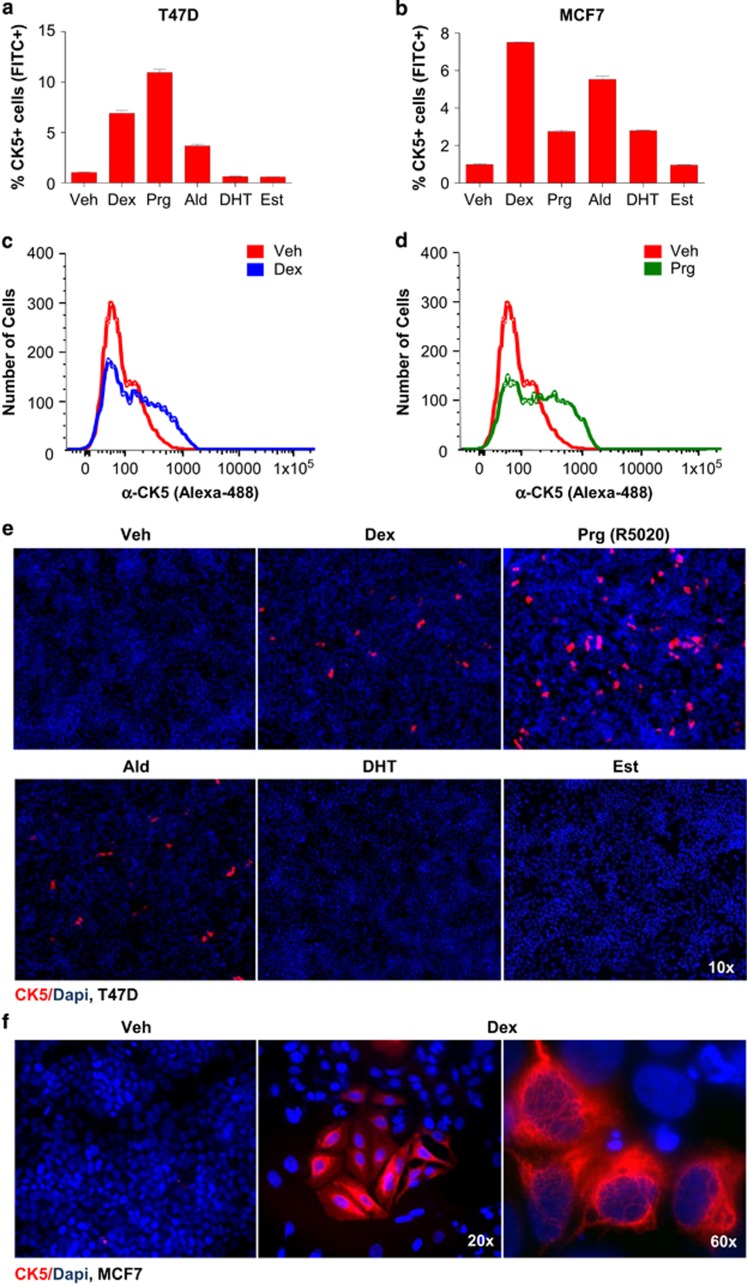
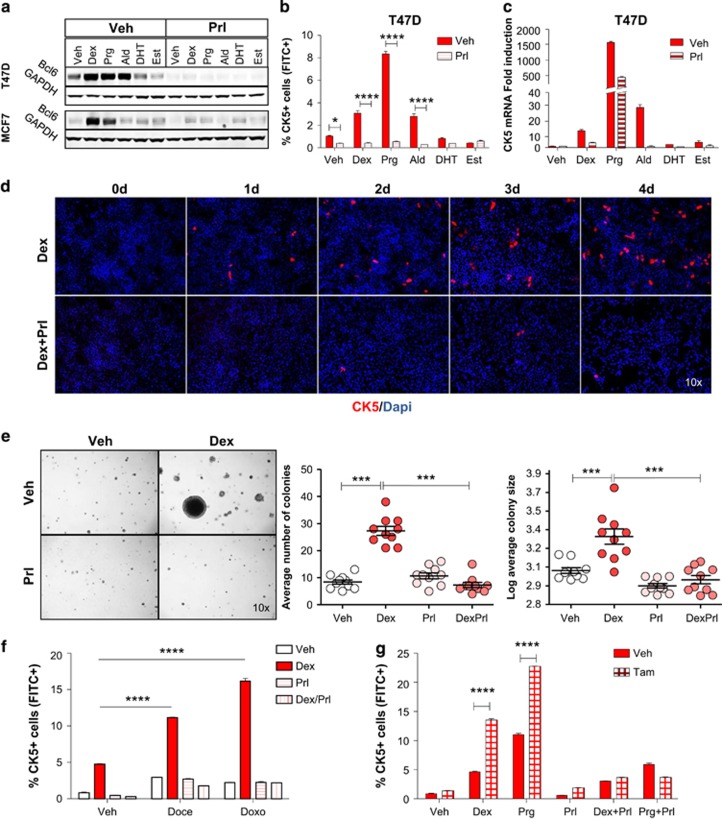
Cultured T47D and MCF7 cell lines were stimulated with cognate ligands for each of the four 3-ketosteroid receptor members, including dexamethasone (Dex), aldosterone (Ald), dihydrotestosterone (DHT) or Prg R5020, with 17β-estradiol (Est) and vehicle included as negative controls. Dex and Ald increased the CK5+ cell population in T47D and MCF7 cells four- to sevenfold as determined by flow cytometry (one-way analysis of variance (ANOVA), P<0.0001 for both analyses; Figure 1a–d). Prg induced CK5+ cells in T47D and MCF7 cell lines 12- and threefold, respectively. DHT induced CK5+ cells threefold in MCF7 cells but less consistently in T47D cells.
Figure 1.
3-ketosteroids induce a CK5+ cell population in ERα-positive breast cancer cell lines. Flow cytometry of CK5+ cells following exposure to 3-ketosteroids or estrogen for 6 days in T47D (a) and MCF7 (b) cell lines. Representative frequency plots of cellular CK5-fluorescence by flow cytometry in T47D cells treated with dexamethasone (Dex; c) or progestin (Prg; d) compared with vehicle (Veh). Representative images of fluorescence-based immunocytochemistry of T47D (e) and MCF7 (f) cell cultures following exposure to 3-ketosteroids for 6 days and stained for CK5 (red) and DAPI (blue).
Immunocytochemistry validated the flow cytometry data, as Dex, Ald and Prg treatment markedly induced CK5+ cells in T47D cells, whereas DHT or Est did not (Figure 1e), and Dex upregulated CK5+ cells in MCF7 cells (Figure 1f). Induction of CK5+ cells was positively associated with hormone receptor expression levels, with Dex most efficacious in high glucocorticoid receptor (GR)-expressing MCF7 cells and Prg most efficacious in high PR-expressing T47D cells (Supplementary Figure S1). Dex-induced CK5+ cells in both T47D and MCF7 showed saturable dose responses consistent with receptor-mediated effects (Supplementary Figure S2). Blockade of GR and PR by the combined receptor antagonist, mifepristone (RU486), selectively blocked the induction of CK5+ cells by Dex and Prg but not Ald in both cell lines, consistent with nuclear receptor-mediated induction of CK5 (two-way ANOVA, P<0.0001 for both analyses; Supplementary Figure S3). In T47D cultures, CK5+ cells increased over 6 days of Dex treatment and were preferentially expanded in clusters, with clusters containing ⩾4 cells increasing after 5 days (Supplementary Figure S4A–C). Faster induction of CK5+ cells occurred in the more Dex-sensitive MCF7 line (Supplementary Figure S4D), although induction of CK5 transcript levels was also rapid in T47D cells (Supplementary Figure S4E).
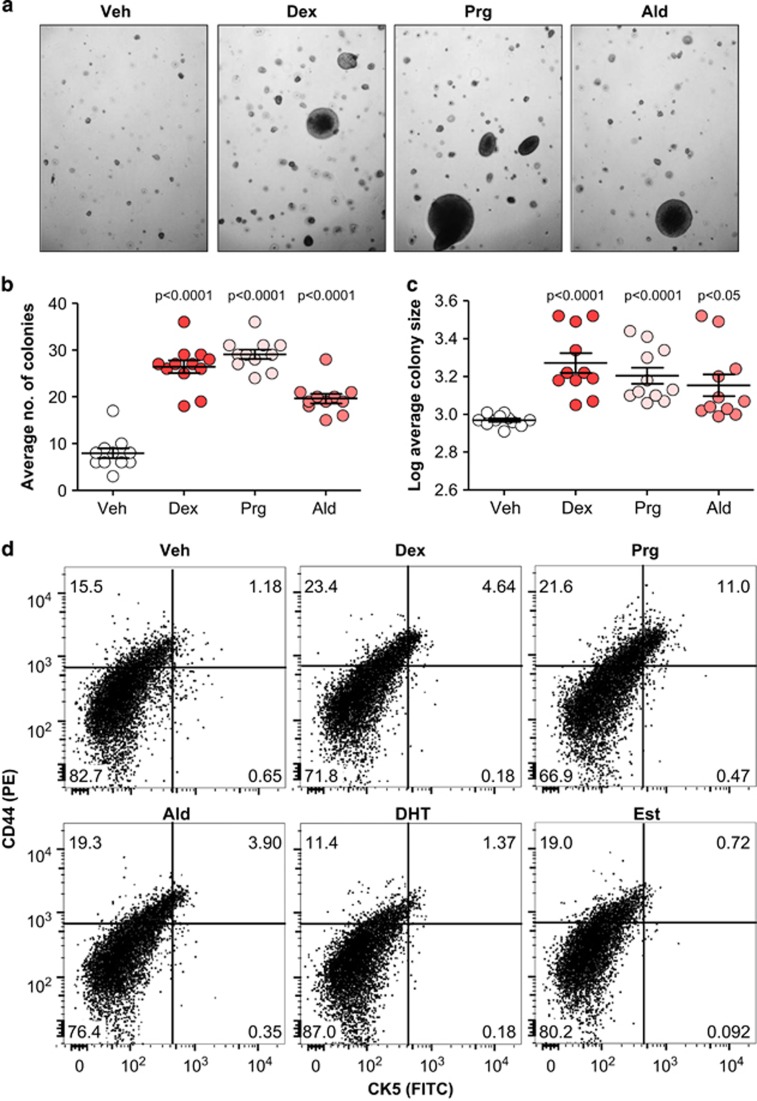
Dex- and Ald-induced CK5+ cells display clonogenic and therapy-resistant properties
To determine whether Dex- and Ald-induced CK5+ cells display stem/progenitor-like features similar to those reported for Prg-induced CK5+ cells, T47D cells pretreated with 3-ketosteroids were seeded in soft agar and colony formation was monitored. Treatment with either steroid induced a two- to threefold increase in colony number (one-way ANOVA, P<0.0001) and a 1.5- to 2-fold increase in colony size (one-way ANOVA, P<0.0001; Figure 2a–c). Previous reports demonstrated that in vivo-derived Prg-induced CK5+ cells could be enriched based on their co-expression of the stem cell marker, CD44.13, 15 Flow cytometry showed that CK5+ cells induced in T47D cell cultures by either Dex, Ald or Prg in vitro co-expressed CD44, with CK5+/CD44+ cells constituting 1.2% of cells in controls and 4.6, 11.0, and 3.9% in Dex-, Prg- and Ald-treated cultures, respectively (Figure 2d).
Figure 2.
3-ketosteroid-treated T47D cells grow in soft agar and express stem cell marker CD44. Representative images of T47D cell colonies in soft agar following stimulation with individual 3-ketosteroids for 6 days (a). Scatter dot plot of colony number per field (b) and colony size per field (c). Mean and s.e.m. are indicated. Multilaser flow cytometry showing CK5 and CD44 expression in T47D cells stimulated with 3-ketosteroids or β-estradiol for 6 days (d).
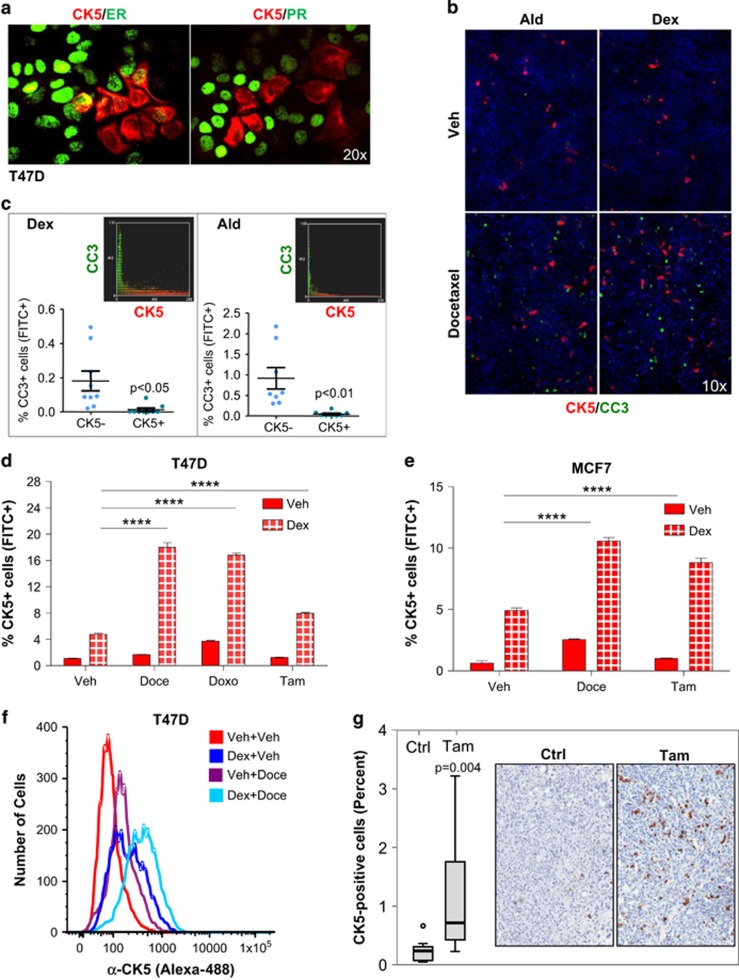
Immunocytochemistry further showed that Dex-induced CK5+ breast cancer cells generally lack ERα and PR expression (Figure 3a), similar to Prg-induced CK5+ cells.13 Dex- or Ald-induced CK5+ cells were resistant to apoptosis as revealed by co-immunocytochemistry for CK5 and cleaved caspase-3 following pretreatment with Dex or Ald for 3 days and subsequent co-treatment with docetaxel (Doce) for 3 days. Image analysis of co-stained cultures demonstrated that cleaved caspase-3-positive cells (green) were significantly less likely to be CK5+ (red) in both Dex-treated (0.18% vs 0.01% Paired samples t-test, P=0.011) and Ald-treated (0.91% vs 0.05% P=0.005) cultures with virtually non-overlapping green and red pixels (Figure 3b and c).
Figure 3.
Dexamethasone- and aldosterone-induced CK5+ cells are resistant to therapy. Representative immunocytochemistry images of T47D cells stained for CK5 (red) and ERα or PR (green) after treatment with Dex for 4 days (a). Note the loss of ERα and PR in CK5+ cells. Representative Immunocytochemistry images (b) and quantification (c) of T47D cells following stimulation with Dex or Ald for 4 days stained for CK5 (red) and cleaved caspase-3 (green). Insets in c show the general non-overlapping CK5 and CC3 staining by scatter plot. Flow cytometry data of CK5+ cell induction following pretreatment with Dex or Veh for 3 days with subsequent addition of Veh, Doce, Doxo or Tam for 3 days in T47D (d) and MCF7 cells (e). Frequency plots of CK5-fluorescence of MCF7 cells following pretreatment with Dex or Veh for 3 days with subsequent addition of Veh or Doce for 3 days (f). Box plots of percent CK5+ cells and representative immunohistochemistry images of T47D xenografts from control- and tamoxifen-treated mice (g). ****P<0.0001.
If Dex-induced CK5+ cells are less sensitive to drug-induced apoptosis, we expected that ERα+ cell lines pretreated with Dex, followed by Doce, doxorubicin or tamoxifen (Tam), would become enriched for CK5+ cells. Indeed, Dex-induced CK5+ cells were further enriched two- to threefold in T47D or MCF7 cell cultures following exposure to such adjuvant drugs (two-way ANOVA, P<0.0001 for both cell lines; Figure 3d–f). Drug-induced expansion of the CK5+ cell population was also demonstrated in vivo as T47D xenograft tumors developed Tam resistance and showed marked enrichment of CK5+ cells relative to control tumors (P=0.004; Figure 3g; Supplementary Figure S5). Collectively, we conclude that Dex- and Ald-induced CK5+ cells display reduced sensitivity to adjuvant agents, similar to that previously observed for Prg-induced CK5+ cells.
3-ketosteroid-induced CK5 expression in ERα+ breast cancer requires BCL6
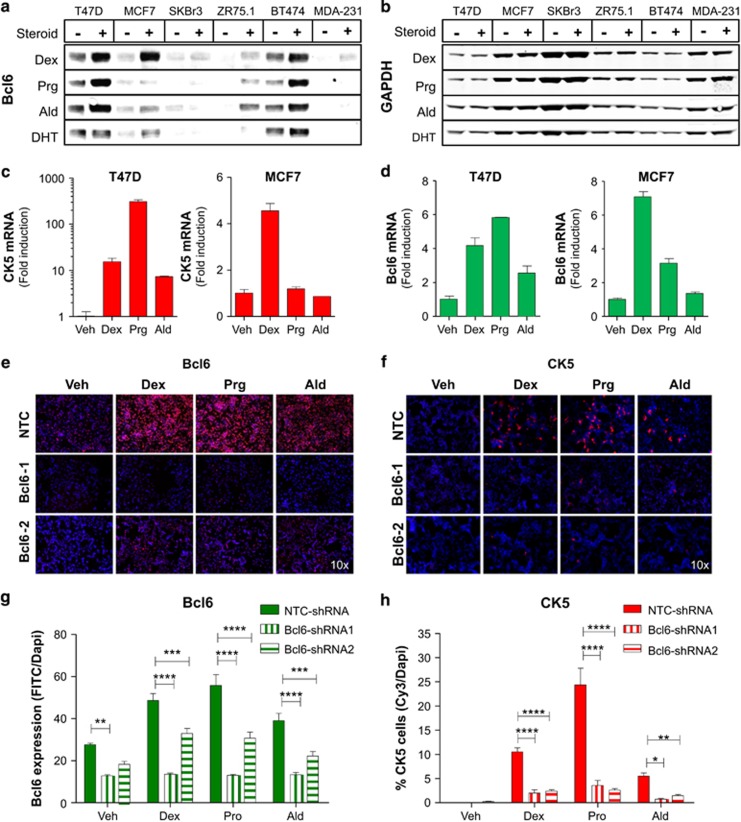
We recesntly reported that Prgs upregulate BCL6 expression in several ERα+ breast cancer cell lines and that induction of BCL6 was required for Prg-induction of CK5+ cells.16 As observed for Prg, western blots of whole-cell lysates revealed that BCL6 protein was frequently upregulated by Dex, Ald and DHT, but not Est, in ERα+ cell lines T47D, MCF7, BT474 and ZR75.1, but not in ERα− cell lines SKBr3 or MDA-MB-231 (Figure 4a and b). BCL6 induction by Dex in MCF7 cells was detected at doses as low as 30 nm and persisted for at least 48 h (Supplementary Figure S6A and B). Dex induction of BCL6 preceded induction of CK5, with maximal levels of BCL6 detected within 8 h vs 5 days for maximal emergence of CK5+ cells in the MCF7 cell line (Supplementary Figure S6B and C). Quantitative reverse transcription PCR experiments demonstrated a positive association between induction of BCL6 and CK5 transcripts by individual 3-ketosteroids in both cell lines, with Dex being most efficacious in MCF7 and Prg in T47D cells (Figure 4c and d). To determine whether 3-ketosteroids generally require BCL6 to induce the CK5+ cell population, two independent small hairpin RNAs in lentiviral vectors were used to knockdown expression of both basal and 3-ketosteroid-induced levels of BCL6 (two-way ANOVA, P<0.0001; Figure 4e and f, Supplementary Figure S7). BCL6 knockdown abolished induction of CK5+ cells by Dex, Ald or Prg as determined by immunocytochemistry (two-way ANOVA, P<0.0001; Figure 4g and h).
Figure 4.
3-ketosteroid-induced CK5 expression in luminal breast cancer requires BCL6. Immunoblots showing BCL6 (a) and GAPDH (b) protein expression in breast cancer cell lines with and without exposure to 3-ketosteroids for 48 h. qRT-PCR analysis of CK5 (c) and BCL6 mRNA (d) extracted from T47D (left) and MCF7 (right) cell lines following treatment with 3-ketosteroids for 48 h. Representative images of immunocytochemistry performed on T47D cells stimulated with 3-ketosteroids for 3 days following exposure to lentiviral-delivered shRNA against NTC or two independent BCL6 shRNAs for 60 h and stained for (e) BCL6 (red) and DAPI. Mean intensity of BCL6 protein expression in each field±s.e.m. (f). Representative images of immunocytochemistry performed on T47D cells stimulated with 3-ketosteroids for 3 days following exposure to lentiviral-delivered shRNA against NTC or two independent BCL6 shRNAs for 60 h and stained for CK5 (red) and DAPI (g), with quantification of data expressed as percent area of CK5+ cells in each field±s.e.m. (h). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
As an independent strategy to test requirement of BCL6 for 3-ketosteroid induction of CK5, we used prolactin to suppress BCL6 protein expression. Our lab and others have demonstrated that prolactin is capable of suppressing basal24, 25 and Prg-induced levels of BCL6 protein in T47D cells.16 Co-treatment of cultured cells with prolactin effectively blocked induction of BCL6 by Dex and Ald as well as Prg in T47D and MCF7 cells (Figure 5a) and Dex-induced BCL6 in ZR75.1 cells (Supplementary Figure S7A). Prolactin suppression of BCL6 displayed sensitive dose responses (Supplementary Figure S7B) and a rapid time response with complete absence of both basal and 3-ketosteroid-induced BCL6 protein within 8 h (Supplementary Figure S7C). Prolactin remained an effective blocker of 3-ketosteroid induction of BCL6 protein even at high steroid concentrations (Supplementary Figure S7D). Consistent with a key role for BCL6 in 3-ketosteroid induction of CK5, prolactin co-treatment effectively blocked induction of CK5 protein and mRNA in T47D cells (two-way ANOVA, P<0.0001; Figure 5b–d; Supplementary Figure S8A), and abolished Dex-induced colony formation in soft agar (one-way ANOVA, P<0.0001; Figure 5e) as well as enrichment of CK5+ cells following exposure to Doce, doxorubicin or Tam (two-way ANOVA, P<0.0001; Figure 5f and g). In contrast, epidermal growth factor co-stimulation was unable to suppress 3-ketosteroid induction of CK5 (Supplementary Figure S8B). Furthermore, neither 3-ketosteroid induction of CK5 nor suppression by prolactin was detected in cultures of the hormone receptor- and prolactin receptor-negative26 MDA-MB-231 breast cancer cell line (Supplementary Figure S8C).
Figure 5.
Prolactin suppresses 3-ketosteroid-induced BCL6 and CK5 expression. Immunoblots showing BCL6 and GAPDH protein expression in T47D and MCF7 cell lines following treatment with 3-ketosteroids with or without prolactin for 6 days (a). Flow cytometry showing CK5+ cell induction (b), qRT-PCR analysis of CK5 mRNA transcripts (c), representative immunocytochemistry of CK5 (red) and DAPI (blue; d) and colony formation on soft agar (e) following treatment with 3-ketosteroids with or without prolactin for 6 days in T47D cells. Flow cytometry showing CK5+ cell induction in T47D cells following 3-ketosteroid treatment with or without concurrent prolactin for 3 days and subsequent addition for 3 days of Doce or Doxo (f) or Tam (g). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Prolactin effectively suppressed 3-ketosteroid induction of cellular BCL6 and CK5 expression in vivo
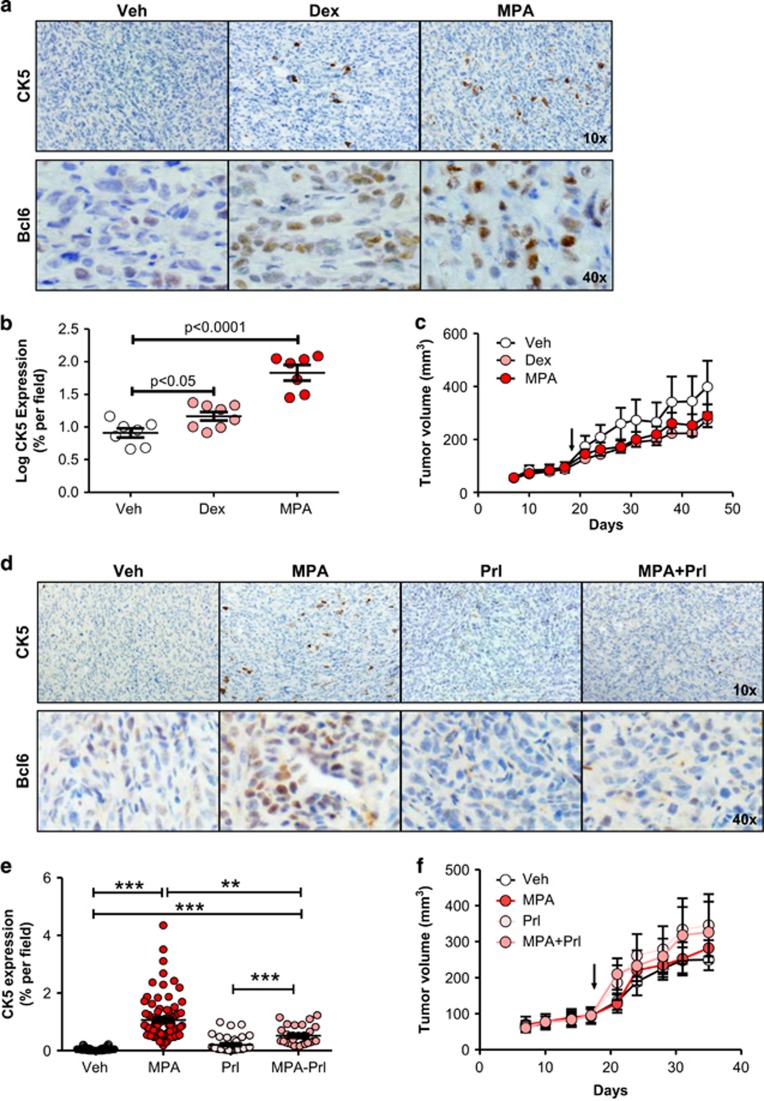
To determine whether our in vitro observations could be recapitulated in vivo, we treated T47D xenograft-bearing mice with vehicle, Dex or medroxyprogesterone acetate (MPA) for 28 days. At the high dose used, MPA broadly activates PR, GR and AR in vivo.27, 28 Immunohistochemical analysis of tumors showed significant induction of a CK5-positive subpopulation in both Dex- and MPA-treated mice compared with controls (linear mixed model, P<0.0001; Figure 6a and b). Furthermore, Dex or MPA treatment for 28 days significantly increased the number of BCL6-positive cells, as well as the intensity of BCL6 protein expression (Figure 6a), while not significantly impacting tumor size (Figure 6c).
Figure 6.
Dexamethasone and medroxyprogesterone acetate treatment induces CK5 and BCL6 expression in vivo, and is suppressed by prolactin. (a) Representative images of T47D xenograft tumors harvested from mice implanted with Veh, Dex or MPA slow-release pellets for 28 days and subjected to immunohistochemistry for CK5 (top) and BCL6 (bottom) using DAB chromogen. (b) Scatter dot plots of the log percent area of CK5+ cells within these tumors. (c) Tumor growth plots in tumor-bearing mice treated with Veh, Dex or MPA as indicated. (d) Representative images of T47D xenograft tumors harvested from mice implanted with slow-release pellets of MPA or Veh with and without co-treatment with biweekly injections of prolactin (Prl) for 21 days and subjected to immunohistochemistry for CK5 (top) and BCL6 (bottom). (e) Scatter dot plots of percent area of CK5+ cells in images taken of these tumors (mean±s.e.m. indicated). (f) Tumor growth plots in mice treated with Veh, MPA, Prl or MPA+Prl as indicated. Arrow indicates initiation of treatment on day 18. *P<0.05; **P<0.01; ***P<0.001.
To determine whether prolactin treatment in vivo would suppress 3-ketosteroid induction of CK5+ cells, mice bearing T47D xenograft tumors were treated with or without high-dose MPA in the presence or absence of human prolactin for 18 days. Prolactin co-treatment effectively suppressed the induction of CK5+ cells by MPA (linear mixed model, P<0.0001) and attenuated MPA-induced levels of BCL6 (Figure 6d and e), consistent with our in vitro observations. Tumor size was not significantly affected (Figure 6f). Prolactin is therefore capable of attenuating 3-ketosteroid induction of BCL6 and CK5 in ERα+ breast cancer cells in vivo.
Quantification of CK5, BCL6 and GR protein expression across subtypes of breast cancer
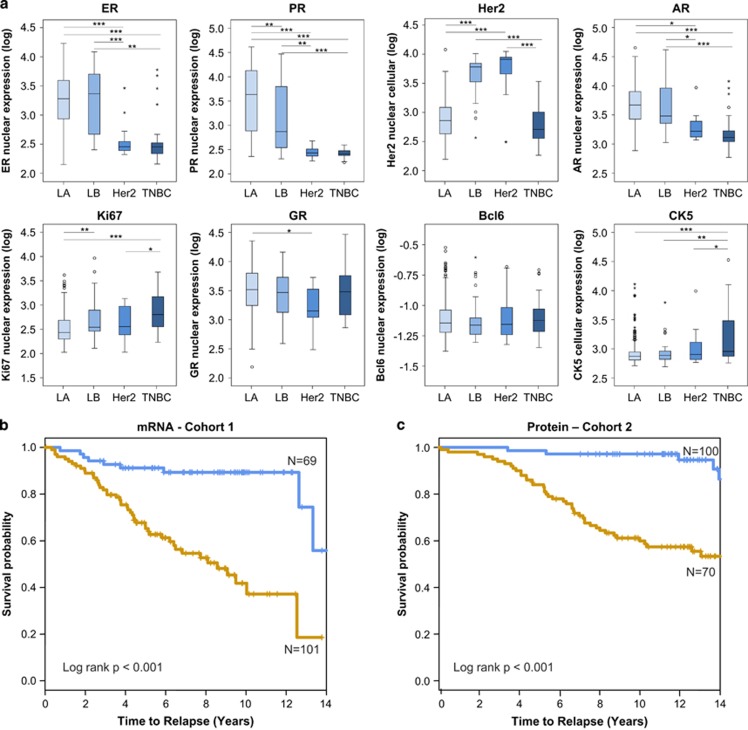
Using quantitative immunofluorescence analysis, we identified a significant number of clinically defined ERα+ breast cancers with elevated levels of CK5 expression. Based on the Tukey Box plot procedure, ~10% of ERα+ primary breast cancer specimens appeared as outliers with elevated levels of CK5 (>75th percentile+1.5 × interquartile range). Interestingly, the proportion of CK5+ outlier cases was similar in tumors classified as Luminal A (immunohistochemistry ERα+/Ki67lo; 10.0% CK5+) and Luminal B (immunohistochemistry ERα+/Ki67hi; 15.4% CK5+). Previous reports indicate that as many as 10–50% of ERα+/PR+ breast cancers display this phenotype of mixed luminal and basal markers.8, 9, 13 As expected, CK5 expression was high in a majority of TNBCs and showed an intermediate range in Her2+ breast cancers. BCL6 was expressed in a significant number of ERα+ breast cancers, although no subtype displayed prominent BCL6 protein levels. Although ERα and PR were expectedly highly expressed in Luminal A and Luminal B tumors, with Her2 highly expressed in Luminal B and Her2+ tumors (one-way ANOVA, P<0.0001 for ERα, PR and Her2 analyses), GR was expressed in Luminal A and Luminal B subtypes as well as the TNBC subtype, with modestly lower levels in Her2-overexpressing tumors (least significant difference test, P<0.05; Figure 7a, Supplementary Figure S9A). Consistent with previous reports, AR was expressed in the ERα+ subtypes and to some extent in TNBC and Her2+ subtypes. Ki67 expression was highest in Luminal B and TNBC subtypes and lowest in Luminal A subtype. Importantly, within the subgroup of mixed ERα+/CK5+ breast cancers ERα and PR levels were comparable to those of classical Luminal A and Luminal B subtypes (Supplementary Figure S9B), indicating that reduction in ERα/PR levels is confined to the CK5+ subpopulation of cells within mixed ERα+/CK5+ tumors.
Figure 7.
CK5 and BCL6 expression is associated with poor outcome in ERα+ breast cancer. (a) Box plots depicting quantitative immunofluorescence-based levels of ERα, PR, Her2, AR, GR, Ki67, BCL6 and CK5 proteins in Luminal A (n=185), Luminal B (n=26), Her2-overexpressing (n=16) and triple-negative (TNBC; n=21) subtypes (*P<0.05; **P<0.01; ***P<0.001). Kaplan–Meier curves indicating high (brown line) or low (blue line) risk of recurrence based on recursive partitioning analysis of CK5 and BCL6 mRNA (b) or CK5 and BCL6 protein (c).
CK5 expression combined with BCL6 expression in node-negative, ERα-positive breast cancer is associated with poor clinical outcome
CK5 expression in TNBC breast cancer defines an aggressive and chemotherapy-resistant subtype,6 but limited information is available on mosaic CK5 expression and clinical outcome in ERα+ breast cancer. As BCL6 expression is associated with CK5 expression, we hypothesized that combined quantitative data on CK5 and BCL6 expression will predict outcome in ERα+ breast cancer. We first used mRNA data from the publicly available Loi data set29 that includes 170 ERα+, lymph node-negative patients, with 85 Tam-treated patients and 85 untreated patients. Remaining known patient characteristics are presented in Supplementary Table 2. Recursive partitioning of cases based on quantitative levels of CK5 and BCL6 transcripts effectively separated favorable and poor prognostic groups (Figure 7b; Supplementary Figure S10A) that remained highly significant after adjustment for other variables in multivariate survival analysis (hazard ratio (HR) 4.34; 95% confidence interval 2.06–18.39; P<0.001; Table 1). Because mRNA levels often correlate poorly with protein levels, we examined BCL6 and CK5 protein levels in an independent cohort of 170 ERα+, lymph node-negative patients, with 50 patients (29.4%) receiving hormone therapy. Recursive partitioning of cases based on quantitative levels of CK5 and BCL6 protein also effectively distinguished favorable and poor prognostic groups (Figure 7c; Supplementary Figure S10B) that remained highly significant after adjustment for other variables in multivariate survival analysis (HR 5.23; 95% confidence interval 1.89–16.37; P=0.004; Table 1). The high-risk group in the mRNA-based analysis is made up of (1) high CK5 expression regardless of BCL6 levels, (2) the combination of low to moderate CK5 with moderate-to-high BCL6 and 3) low CK5 expression regardless of BCL6 levels. A similar pattern was found in protein analyses, with high risk associated with (1) high CK5 expression regardless of BCL6 levels, (2) high BCL6 in the absence of CK5 (l<25%-tile of expression) and (3) the combination of low CK5 with low-to-moderate BCL6. Although ERα+ breast cancer with high CK5 and/or high BCL6 intuitively fit a mixed ERα+/CK5+ profile, the poor outcome in the fraction of ERα+ tumors with the very lowest CK5 levels most likely reflects the Luminal B subclass.
Table 1. Multivariable analysis.
|
mRNA cohort (n=170) |
Protein cohort (n=170) |
||||
|---|---|---|---|---|---|
| AHRa | P-value | AHR | P-value | ||
| Age | 0.98 (0.95, 1.02) | 0.89 | Age | 1.03 (1.00, 1.13) | 0.03 |
| Tumor size | 1.63 (1.11, 2.63) | 0.01 | — | ||
| Endocrine therapy | Endocrine therapy | ||||
| No | — | No | — | ||
| Yes | 0.86 (0.35, 1.43) | 0.69 | Yes | 0.68 (0.18, 2.76) | 0.69 |
| Grade | AJCC stage | ||||
| 2 | 0.77 (0.40, 1.67) | 0.75 | 1 | — | |
| 3 | 1.40 (0.49, 5.39) | 0.19 | 2 | 1.64 (0.48, 3.48) | 0.21 |
| CK5/BCL6-related risk | CK5/BCL6-related risk | ||||
| Low | — | Low | — | ||
| High | 4.34 (2.06, 18.39) | <0.001 | High | 5.23 (1.89, 16.37) | 0.004 |
Adjusted hazard ratio.
Discussion
The present study documents a previously unrecognized ability of glucocorticoids and other 3-ketosteroids to expand a therapy-resistant CK5+ cell population in breast cancer of the mixed ERα+/CK5+ subclass. This represents a paradigm shift in terms of the range of hormonal and pharmacological impact on mixed ERα+/CK5+ breast cancer beyond what was previously thought to be restricted to Prgs.19 The discovery of glucocorticoid-driven expansion of the CK5+ cell population in mixed ERα+/CK5+ breast cancer is of particular clinical relevance considering (1) the widespread pharmacological use of glucocorticoids for common breast cancer comorbidities, (2) the standard administration of glucocorticoids to breast cancer patients who receive adjuvant chemotherapy and (3) stress-related elevation of endocrine glucocorticoids associated with a breast cancer diagnosis. Glucocorticoids have been implicated in resistance to chemotherapy in breast cancer as well as other solid tumors,30, 31, 32 and stress-related elevation of endocrine glucocorticoids is associated with unfavorable clinical outcome in breast cancer patients.33, 34, 35, 36 Although glucocorticoids may promote therapy resistance through multiple mechanisms, glucocorticoid-driven expansion of a de-differentiated and hormone receptor-negative CK5+ cell population may be particularly important for mixed ERα+/CK5+ breast cancer. As mineralocorticoid and androgen receptors are expressed in normal and malignant breast epithelia,37, 38 further investigations of these 3-ketosteroids are warranted. Interestingly, a randomized Phase III study of ERα+ breast cancer patients on Tam revealed that co-treatment with the broad 3-ketosteroid MPA significantly worsened outcome.39 Additional clinical significance for a 3-ketosteroid receptor-BCL6-CK5 pathway was provided by the discovery of a strong association between poor clinical outcome and elevated CK5/BCL6 expression in ERα+ breast cancer, associations that persisted in multivariate analyses with adjusted HRs much greater than those observed in limited reports of CK5 expression alone.7, 9, 16 Thus, combining quantitative expression levels of CK5 and BCL6 may have diagnostic utility for ERα+ breast cancer.
Glucocorticoid-mediated effects on the CK5+ cell population were consistent with that seen following Prg stimulation for all readouts examined. The Dex-expanded CK5+ cell population was characterized by enrichment of stem cell marker CD44, loss of ERα expression, enhanced colony-forming ability in soft agar and broad therapy resistance against several classes of drugs, including the DNA-damaging agent doxorubicin, the microtubule disrupting agent Doce, and the selective ER modifier Tam. GR activation in mixed ERα+/CK5+ breast cancer also replicated Prg induction of oncoprotein and transcriptional repressor BCL640, 41, 42 prior to expansion of the CK5+ cell population. Upregulation of BCL6 was required for CK5+ cell expansion by glucocorticoids, Ald and Prgs based on BCL6 knockdown and effective downregulation of BCL6 protein by prolactin. The results were validated in vivo by the demonstration that T47D xenograft tumors grown in 3-ketosteroid-treated mice displayed markedly elevated BCL6 protein accompanied by expansion of the CK5+ cell population, effects that were blocked by prolactin. Our data therefore indicate that the 3-ketosteroid nuclear receptor family shares a common mechanism for CK5+ cell induction in mixed ERα+/CK5+ breast cancer that involves upregulation of BCL6. Future studies will investigate three predicted glucocorticoid response elements (UCSC Genome Browser) in the BCL6 gene promoter and five in the CK5 gene promoter that may mediate 3-ketosteroid-induced expansion of CK5+ cells. It should be noted that 3-ketosteroids induce BCL6 more broadly throughout the cancer cell population than CK5, which becomes expressed only in a subpopulation of cells. We hypothesize that BCL6-mediated upregulation of CK5 expression involves yet-to-be identified molecular intermediaries that act on a select subpopulation of responsive cells. Two alternative but not mutually exclusive possibilities are that (1) a subset of cells intrinsically respond to BCL6 activation with CK5 upregulation, and (2) BCL6 induces paracrine factors to which a subset of cells respond with CK5 upregulation. BCL6 is already implicated in the promotion of breast cancer, as BCL6 attenuated differentiation and blocked apoptosis in mouse mammary EpH4 cells,43 and BCL6 is elevated in high-grade and metastatic breast cancers.24, 25, 44 Follow-up studies will determine whether BCL6 upregulation alone is sufficient for expansion of the CK5+ cell population in tumors of the mixed ER+/CK5+ subtype, and will seek to extend the mechanistic studies from laboratory cell lines to patient-derived xenograft models and murine genetic models.
The efficacy of prolactin to abrogate expansion of the de-differentiated CK5+ cell population induced by 3-ketosteroids or chemo- and anti-estrogen therapies in ERα+ breast cancer is intriguing, but is consistent with the established role of prolactin in the maintenance of mammary epithelial differentiation.45, 46, 47, 48 Loss of prolactin signaling may potentiate the emergence of this CK5+/ERα− cell population in a subgroup of ERα+ breast cancers through unopposed BCL6 activity and lead to therapy resistance. Loss of signaling by signal transducer and activator of transcription-5 (STAT5), a key mediator of prolactin signaling, is associated with poor clinical outcome and increased risk of failure of anti-estrogen therapy.45, 46, 49, 50 Although prolactin receptor expression is typically maintained or increased in breast cancer51 and may be further boosted by co-recruitment of Jak1 in addition to Jak2,26 disruptive mechanisms underlying observed frequent loss of prolactin receptor signaling include extracellular acidosis,52 elevated levels of the Jak2 phosphatase PTP1B53 or loss of STAT5a protein expression or phosphorylation.50 Suppression of prolactin signaling is potentially of particular relevance for mixed ERα+/CK5+ breast cancer. These observations shed new light on evidence that prolactin–STAT5 signaling inhibits invasive features and maintains cell differentiation of some ERα+ breast cancer lines.48, 54
In conclusion, the new data broadly implicate receptors for 3-ketosteroids as drivers of expansion of the therapy-resistant CK5+ cell population in mixed ERα+/CK5+ breast cancer. A common mechanism involves upregulation of the transcriptional repressor BCL6. The progress supports the concept that mixed ERα+/CK5+ breast cancers develop resistance to anti-estrogens and chemotherapies driven by selective expansion of a CK5+/ERα− subpopulation of cells. The observations provide rationale for exploring 3-ketosteroid receptor antagonists or BCL6 inhibitors in patients with mixed ERα+/CK5+ breast cancer. Conversely, progression of mixed ERα+/CK5+ breast cancer may be promoted by pharmacological agonists of 3-ketosteroid receptors or physiological or pathophysiological conditions (for example, stress, pregnancy and endocrinopathies) that elevate 3-ketosteroids. Finally, loss of prolactin–STAT5 signaling may directly contribute to progression of mixed ERα+/CK5+ breast cancer.
Materials and methods
Cell culture
Cell lines were cultured as previously described.16, 55 Media were changed to RPMI+5% charcoal-stripped fetal calf serum (Thermo Scientific, Philadelphia, PA, USA) 24 h before experiments. Cells were treated with 20 nm recombinant human prolactin (AFP795; National Hormone and Peptide Program, Torrance, CA, USA), 1–10 μm Dex, 100 nm Ald, 10 nm DHT, 1 nm 17β-Est, 10 nm epidermal growth factor (Sigma, St Louis, MO, USA), 20 nm R5020 (PerkinElmer, Waltham, MA, USA), and/or 1 μm RU486 (Sigma) as indicated with fresh media every 3 days. Dex was applied at 1 μm for MCF7 and 10 μm for T47D, as determined by dose response experiments. For adjuvant therapy experiments, cells were treated with hormones for 3 days before treatment with 10 nm Doce, 1 μm doxorubicin, or 1 nm 17β-Est+1 nM 4-Hydroxytamoxifen (Sigma) for 3 days. Cell lines were obtained from ATCC (authenticated by Short Tandem Repeat profiling and confirmed mycoplasma-negative) and cultured for <6 months.
Lentiviral infection
Lentivirus was produced as described previously.16 Cells were infected with lentivirus overnight and allowed to grow for 48 h before hormone induction for an additional 6 days.
Quantitative reverse transcription PCR
Quantitative reverse transcription PCR assays were performed as described previously16, 24 using corresponding primers (Supplementary Table S1).
Immunoblotting
Performed as described previously.53 Proteins were immunoblotted with antibodies to phospho-STAT5 (#611964; BD Biosciences, East Rutherford, NJ, USA), total STAT5 (#610192; BD Biosciences), BCL6 (#M7211; Dako, Carpinteria, CA, USA), CK5 (#MS-1896-S; Thermo Scientific) or GAPDH (sc-25778; Santa Cruz Biotechnology, Dallas, TX, USA), followed by AlexaFluor-680-conjugated goat anti-mouse IgG (Invitrogen, Carlsbad, CA, USA) or IRDye-800-CW-conjugated goat anti-rabbit IgG (Licor, Lincoln, NE, USA). Immunoblots were scanned using Odyssey Infrared Imaging System (Licor).
Immunocytochemistry
Performed as described previously.16 Primary antibodies to BCL6, CK5, ERα (EP1; Dako), PR-B (#3157S; Cell Signaling, Beverly, MA, USA), GR (#3660; Cell Signaling), GATA3 (#5852S; Cell Signaling) or Cleaved Caspase-3 (#9661; Cell Signaling) were applied, followed by AlexaFluor-555-conjugated goat anti-mouse/488-conjugated goat anti-rabbit antibodies (Invitrogen). Images were captured using the OLY-100 microscope.
Flow cytometry
Cells were collected, fixed, permeabilized and stained with FITC-conjugated CK5 antibody (sc-32721; Santa Cruz Biotechnology) and/or PE-conjugated CD44 (#562818; BD Biosciences), run on a BD LSRII instrument, and analyzed by FlowJo (Ashland, OR, USA). The CK5+ gate was defined for 1% of vehicle cells as positive and applied to each specimen.
Soft agar assay
T47D cells were pretreated for 6 days and allowed to form colonies in soft agar for 3 weeks. 10 images/well were analyzed using ImageJ software (NIH, Bethesda, MD, USA) for number and mean size of colonies >600 pixels.
Xenograft studies
T47D xenotransplants were performed in immunodeficient estrogenized nude female mice (8–10 weeks old, 17β-Est pellets; 0.72 mg/90 days release; IRA, Sarasota, FL, USA) as described previously.24 For the Tam study, tumors growing in control-treated (n=5) or Tam-treated mice (n=5; 10 mg/90 days release; IRA) were harvested as indicated. For the Dex/MPA studies, mice (n=21) were randomized and implanted with Dex (1.5 mg/60 days) or MPA (10 mg/60 days) subcutaneous pellets. Specified randomized animals (n=6) were injected subcutaneously with 4 μg/g body mass of human prolactin biweekly for 3 weeks. Tumors from animals that died were excluded from the analysis. All animal studies were approved by Thomas Jefferson University's IACUC.
Immunohistochemistry
Performed as described previously.16, 24 Tumor sections were stained with CK5 and BCL6, and images were taken at 20–40 × magnification using the Olympus BX40 microscope and analyzed for percent area of CK5+ cells in a blinded manner. A breast cancer tissue microarray representing an unselected cohort of patients from Thomas Jefferson University Hospital pathology archives under Institutional Review Board-approved protocols was evaluated for nuclear or cytoplasmic BCL6, CK5, ER (#M7047; Dako), AR (#MDR2044; Molecular Detection and Retrieval, Windber, PA, USA), PR (#M3569; Dako), Her2 (#A0485; Dako) and GR immunofluorescent staining on 276 cases using an Autostainer Plus (Dako). High-resolution digital images were captured at 20 × using Scanscope FL (Aperio, Sausalito, FL, USA). Quantitative analyses for average staining intensity were performed using Tissue Studio digital pathology image analysis software (Definiens, Carlsbad, CA, USA).
Statistical methods
Appropriate statistical analyses, post hoc tests and Bartlett's test for equal variance were performed using R (R Foundation for Statistical Computing, Vienna, Austria), SPSS (IBM, Armonk, NY, USA) and GraphPad Prism software (Graphpad Software, La Jolla, CA, USA). All in vitro experiments presented are representative and were replicated at least two times. Data presented in bar graphs represent mean (n=3)±s.e.m. Sample sizes for relevant experiments were determined by power analyses conducted during experiment planning. CK5 and BCL6 mRNA expression data and clinical annotations from Loi et al.29 were downloaded from the GeneChip Oncology Database, which contained Affymetrix HG-U133A expression data for ERα+, lymph node-negative breast cancers with focus on relapse-free survival and data from the BCL6 (X203140_at, X215990_s_at) and CK5 (X201820_at) probes. A breast cancer array with clinical annotation from Thomas Jefferson University Hospital pathology archives focusing on ERα+, lymph node-negative patients was developed and protein expression of BCL6 and CK5 was quantified from immunofluorescent staining analyzed by Tissue Studio (Definiens Inc, Munich, Germany). For each cohort, a recursive partitioning analysis56 of BCL6 and CK5 expression data were performed to determine marker combinations informative of relapse-free survival, which was then incorporated in a multivariable Cox model controlled for age at diagnosis, hormone therapy status, grade and tumor size in the mRNA model, and age at diagnosis, hormone therapy status and stage in the protein model. Bootstrap was completed around the recursive partitioning and Cox modeling process. Confidence intervals and P-values for adjusted HRs are reported based on 2000 bootstrap samples. The R packages, rpart, survival and bootstrap were used to complete survival analysis.
Acknowledgments
This work is supported by grants from Susan G Komen (KG091116), Ladies of Port Richmond and NCI (CA185918, CA188575; Support Grant 1P30CA56036), and in part, under a Commonwealth University Research Enhancement Program grant with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. We thank Colleen Lord for expert editorial assistance.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Anderson WF, Chatterjee N, Ershler WB, Brawley OW. Estrogen receptor breast cancer phenotypes in the Surveillance, Epidemiology, and End Results database. Breast Cancer Res Treat 2002; 76: 27–36. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Ann Rev Med 2011; 62: 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol 2007; 608: 1–22. [DOI] [PubMed] [Google Scholar]
- Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol 2010; 28: 1684–1691. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature 2001; 414: 105–111. [DOI] [PubMed] [Google Scholar]
- Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 2008; 14: 1368–1376. [DOI] [PubMed] [Google Scholar]
- Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF et al. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol 2004; 203: 661–671. [DOI] [PubMed] [Google Scholar]
- Haughian JM, Pinto MP, Harrell JC, Bliesner BS, Joensuu KM, Dye WW et al. Maintenance of hormone responsiveness in luminal breast cancers by suppression of Notch. Proc Natl Acad Sci USA 2012; 109: 2742–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensuu K, Leidenius M, Kero M, Andersson LC, Horwitz KB, Heikkila P. ER, PR, HER2, Ki-67 and CK5 in Early and Late Relapsing Breast Cancer-Reduced CK5 Expression in Metastases. Breast Cancer 2013; 7: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox AJ, Scaling AL, Pinto MP, Bliesner BS, Haughian JM, Abdel-Hafiz HA et al. Modeling Luminal breast cancer heterogeneity: combination therapy to suppress a hormone receptor-negative, cytokeratin 5-positive subpopulation in Luminal disease. Breast Cancer Res 2014; 16: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes J, Lopes N, Milanezi F, Schmitt FC. P-cadherin and cytokeratin 5: useful adjunct markers to distinguish basal-like ductal carcinomas in situ. Virchows Arch 2007; 450: 73–80. [DOI] [PubMed] [Google Scholar]
- Rakha EA, El-Sayed ME, Green AR, Paish EC, Lee AH, Ellis IO. Breast carcinoma with basal differentiation: a proposal for pathology definition based on basal cytokeratin expression. Histopathology 2007; 50: 434–438. [DOI] [PubMed] [Google Scholar]
- Kabos P, Haughian JM, Wang X, Dye WW, Finlayson C, Elias A et al. Cytokeratin 5 positive cells represent a steroid receptor negative and therapy resistant subpopulation in luminal breast cancers. Breast Cancer Res Treat 2011; 128: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorius CA, Harvell DM, Shen T, Horwitz KB. Progestins initiate a luminal to myoepithelial switch in estrogen-dependent human breast tumors without altering growth. Cancer Res 2005; 65: 9779–9788. [DOI] [PubMed] [Google Scholar]
- Horwitz KB, Dye WW, Harrell JC, Kabos P, Sartorius CA. Rare steroid receptor-negative basal-like tumorigenic cells in luminal subtype human breast cancer xenografts. Proc Natl Acad Sci USA 2008; 105: 5774–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Tran TH, Peck AR, Girondo MA, Liu C, Goodman CR et al. Prolactin suppresses a progestin-induced CK5-positive cell population in luminal breast cancer through inhibition of progestin-driven BCL6 expression. Oncogene 2014; 33: 2215–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BH, Axlund SD, Kabos P, Reid BG, Schaack J, Sartorius CA et al. A high-content assay to identify small-molecule modulators of a cancer stem cell population in luminal breast cancer. J Biomol Screen 2012; 17: 1211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cittelly DM, Finlay-Schultz J, Howe EN, Spoelstra NS, Axlund SD, Hendricks P et al. Progestin suppression of miR-29 potentiates dedifferentiation of breast cancer cells via KLF4. Oncogene 2013; 32: 2555–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axlund SD, Yoo BH, Rosen RB, Schaack J, Kabos P, Labarbera DV et al. Progesterone-inducible cytokeratin 5-positive cells in luminal breast cancer exhibit progenitor properties. Horm Cancer 2013; 4: 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CC, Hendy SC, Shukin RJ, Cheng H, Bruchovsky N, Koop BF et al. Determinants of DNA sequence specificity of the androgen, progesterone, and glucocorticoid receptors: evidence for differential steroid receptor response elements. Mol Endocrinol 1999; 13: 2090–2107. [DOI] [PubMed] [Google Scholar]
- Wan Y, Nordeen SK. Overlapping but distinct gene regulation profiles by glucocorticoids and progestins in human breast cancer cells. Mol Endocrinol 2002; 16: 1204–1214. [DOI] [PubMed] [Google Scholar]
- Li X, Wong J, Tsai SY, Tsai MJ, O'Malley BW. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol Cell Biol 2003; 23: 3763–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Roqueiro D, Huang L, Owen JK, Xie A, Navarro A et al. Genome-wide progesterone receptor binding: cell type-specific and shared mechanisms in T47D breast cancer cells and primary leiomyoma cells. PLoS one 2012; 7: e29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Utama FE, Lin J, Yang N, Sjolund AB, Ryder A et al. Prolactin inhibits BCL6 expression in breast cancer through a Stat5a-dependent mechanism. Cancer Res 2010; 70: 1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene 2007; 26: 224–233. [DOI] [PubMed] [Google Scholar]
- Neilson LM, Zhu J, Xie J, Malabarba MG, Sakamoto K, Wagner KU et al. Coactivation of janus tyrosine kinase (Jak)1 positively modulates prolactin-Jak2 signaling in breast cancer: recruitment of ERK and signal transducer and activator of transcription (Stat)3 and enhancement of Akt and Stat5a/b pathways. Mol Endocrinol 2007; 21: 2218–2232. [DOI] [PubMed] [Google Scholar]
- Ouatas T, Halverson D, Steeg PS. Dexamethasone and medroxyprogesterone acetate elevate Nm23-H1 metastasis suppressor gene expression in metastatic human breast carcinoma cells: new uses for old compounds. Clin Cancer Res 2003; 9: 3763–3772. [PubMed] [Google Scholar]
- Bentel JM, Birrell SN, Pickering MA, Holds DJ, Horsfall DJ, Tilley WD. Androgen receptor agonist activity of the synthetic progestin, medroxyprogesterone acetate, in human breast cancer cells. Mol Cell Endocrinol 1999; 154: 11–20. [DOI] [PubMed] [Google Scholar]
- Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol 2007; 25: 1239–1246. [DOI] [PubMed] [Google Scholar]
- Wu W, Chaudhuri S, Brickley DR, Pang D, Karrison T, Conzen SD. Microarray analysis reveals glucocorticoid-regulated survival genes that are associated with inhibition of apoptosis in breast epithelial cells. Cancer Res 2004; 64: 1757–1764. [DOI] [PubMed] [Google Scholar]
- Pang D, Kocherginsky M, Krausz T, Kim SY, Conzen SD. Dexamethasone decreases xenograft response to Paclitaxel through inhibition of tumor cell apoptosis. Cancer Biol Ther 2006; 5: 933–940. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wenger T, Mattern J, Ilea S, Frey C, Gutwein P et al. Clinical and mechanistic aspects of glucocorticoid-induced chemotherapy resistance in the majority of solid tumors. Cancer Biol Ther 2007; 6: 278–287. [DOI] [PubMed] [Google Scholar]
- Palesh O, Butler LD, Koopman C, Giese-Davis J, Carlson R, Spiegel D. Stress history and breast cancer recurrence. J Psychosom Res 2007; 63: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volden PA, Conzen SD. The influence of glucocorticoid signaling on tumor progression. Brain Behav Immun 2013; 30: S26–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chida Y, Hamer M, Wardle J, Steptoe A. Do stress-related psychosocial factors contribute to cancer incidence and survival? Nat Clin Pract Oncol 2008; 5: 466–475. [DOI] [PubMed] [Google Scholar]
- Costanzo ES, Sood AK, Lutgendorf SK. Biobehavioral influences on cancer progression. Immunol Allergy Clin North Am 2011; 31: 109–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde I, Paniagua R, Fraile B, Lucio J, Arenas MI. Glucocorticoid receptor changes its cellular location with breast cancer development. Histol Histopathol 2008; 23: 77–85. [DOI] [PubMed] [Google Scholar]
- Sasano H, Frost AR, Saitoh R, Matsunaga G, Nagura H, Krozowski ZS et al. Localization of mineralocorticoid receptor and 11 beta-hydroxysteroid dehydrogenase type II in human breast and its disorders. Anticancer Res 1997; 17: 2001–2007. [PubMed] [Google Scholar]
- Byrne MJ, Gebski V, Forbes J, Tattersall MH, Simes RJ, Coates AS et al. Medroxyprogesterone acetate addition or substitution for tamoxifen in advanced tamoxifen-resistant breast cancer: a phase III randomized trial. Australian-New Zealand Breast Cancer Trials Group. J Clin Oncol 1997; 15: 3141–3148. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 2000; 13: 199–212. [DOI] [PubMed] [Google Scholar]
- Wagner SD, Ahearne M, Ferrigno PK. The role of BCL6 in lymphomas and routes to therapy. Br J Haematol 2011; 152: 3–12. [DOI] [PubMed] [Google Scholar]
- Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature 2011; 473: 384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarajah S, Hunter P, Kraman M, Steele D, Lakhani S, Bobrow L et al. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiation. Oncogene 2003; 22: 5572–5578. [DOI] [PubMed] [Google Scholar]
- Wu Q, Liu X, Yan H, He YH, Ye S, Cheng XW et al. B-cell lymphoma 6 protein stimulates oncogenicity of human breast cancer cells. BMC Cancer 2014; 14: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck AR, Witkiewicz AK, Liu C, Stringer GA, Klimowicz AC, Pequignot E et al. Loss of nuclear localized and tyrosine phosphorylated Stat5 in breast cancer predicts poor clinical outcome and increased risk of antiestrogen therapy failure. J Clin Oncol 2011; 29: 2448–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, Nishio M, Ando Y, Zhang Z, Hamaguchi M, Mita K et al. Stat5 expression predicts response to endocrine therapy and improves survival in estrogen receptor-positive breast cancer. Endocr Relat Cancer 2006; 13: 885–893. [DOI] [PubMed] [Google Scholar]
- Sultan AS, Xie J, LeBaron MJ, Ealley EL, Nevalainen MT, Rui H. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene 2005; 24: 746–760. [DOI] [PubMed] [Google Scholar]
- Nouhi Z, Chughtai N, Hartley S, Cocolakis E, Lebrun JJ, Ali S. Defining the role of prolactin as an invasion suppressor hormone in breast cancer cells. Cancer Res 2006; 66: 1824–1832. [DOI] [PubMed] [Google Scholar]
- Nevalainen MT, Xie J, Torhorst J, Bubendorf L, Haas P, Kononen J et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J Clin Oncol 2004; 22: 2053–2060. [DOI] [PubMed] [Google Scholar]
- Peck AR, Witkiewicz AK, Liu C, Klimowicz AC, Stringer GA, Pequignot E et al. Low levels of Stat5a protein in breast cancer are associated with tumor progression and unfavorable clinical outcomes. Breast Cancer Res 2012; 14: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov A, Varghese B, Tran TH, Liu C, Rui H, Fuchs SY. Impaired turnover of prolactin receptor contributes to transformation of human breast cells. Cancer Res 2009; 69: 3165–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Liu C, Peck AR, Girondo MA, Yanac AF, Tran TH et al. Prolactin-Stat5 signaling in breast cancer is potently disrupted by acidosis within the tumor microenvironment. Breast Cancer Res 2013; 15: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KJ, Peck AR, Liu C, Tran TH, Utama FE, Sjolund AB et al. PTP1B suppresses prolactin activation of Stat5 in breast cancer cells. Am J Pathol 2010; 177: 2971–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan AS, Brim H, Sherif ZA. Co-overexpression of Janus kinase 2 and signal transducer and activator of transcription 5a promotes differentiation of mammary cancer cells through reversal of epithelial-mesenchymal transition. Cancer Sci 2008; 99: 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Tran TH, Peck AR, Liu C, Ertel A, Lin J et al. Global profiling of prolactin-modulated transcripts in breast cancer in vivo. Mol Cancer 2013; 12: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Stat 2006; 15: 651–674. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.