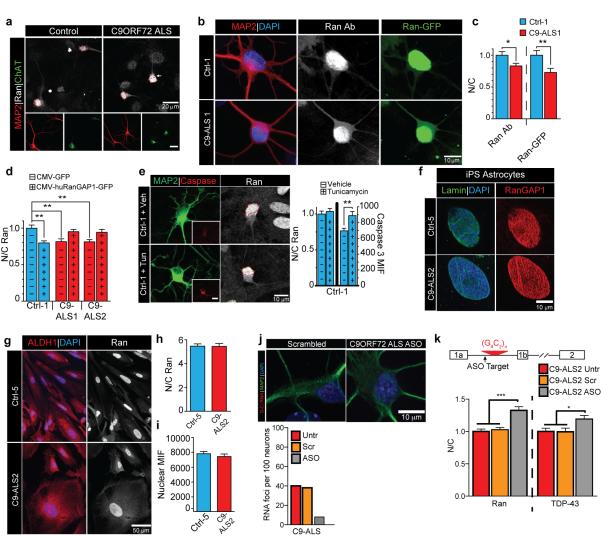
Extended Data Fig. 8. C9ORF72 HRE disrupts the cytoplasmic/nuclear Ran gradient.
(a) Representative images of disrupted N/C Ran gradient in C9-ALS ChAT+ iPS neurons. (b-c) Representative images and quantification of control (top row) or C9-ALS iPS neurons (bottom row) expressing Ran-GFP that are co-stained with Ran and MAP2. Both Ran antibody and Ran-GFP indicate a reduced N/C Ran ratio. (d) Overexpression of RanGAP1-GFP rescues the N/C Ran ratio in C9-ALS iPS neurons. (e) Control iPS neurons treated with Tunicamycin show enhanced level of activated Caspase 3 in the soma but no change in N/C Ran localization compared to controls with vehicle treatment. (f) RanGAP1 is not aggregated in Ctrl and C9-ALS iPS astroglia. (g) Representative image of N/C Ran in C9-ALS astrocytes when identified using the pan astroglial ALDH1 marker. (h) N/C Ran is not altered in C9-ALS astroglia when comparing astrocytes of a similar size. (i) Mean intensity fluorescence (MIF) of nuclear Ran does not differ in ctrl or C9-ALS astroglia. (j) Representative image of C9-ALS iPS neuron with G4C2 RNA foci in approximately 40% of MAP2+ neurons at 50 - 70 DIV. Number of C9-ALS iPS neurons with RNA foci is reduced with C9ORF72 RNA targeting ASOs compared to scrambled/non-targeting ASOs to <10% of iPS neurons. (k) ASOs that reduce G4C2 RNA foci also enhance N/C Ran and N/C TDP-43 ratios. (*, p<0.05; **, p<0.01; ****, p<0.0001).