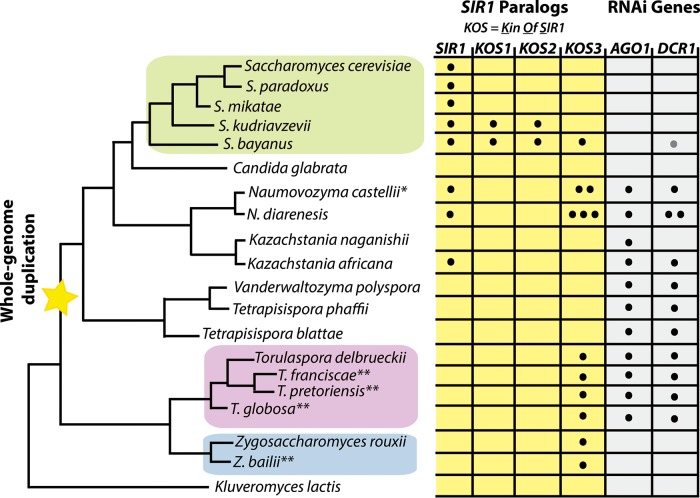
FIG 2.
SIR1 paralogs and RNAi genes in the family Saccharomycetaceae. Depicted is a phylogenetic tree of budding yeast species in the family Saccharomycetaceae, along with the SIR1 paralogs and RNAi gene paralogs (where applicable; some species, e.g., K. lactis, do not have SIR1 or the RNAi genes AGO1 and DCR1). The number of dots within each box indicates the number of copies of that particular paralog in the genome (e.g., N. castellii has two highly similar KOS3 paralogs). S. cerevisiae contains the defining SIR1 gene, whereas S. bayanus contains four SIR1 genes: SIR1 and three KOS paralogs. KOS3 is the earliest SIR1 paralog, deduced to have originated prior to the whole-genome duplication. T. delbrueckii has the budding yeast orthologs of AGO1 and DCR1. All sequenced species in the Zygosaccharomyces and Torulaspora clades have a KOS3 paralog in their genomes. *, N. castellii also has a fourth SIR1 paralog, KOS4, specific to that species (not shown for simplicity); **, results from additional species (Zygosaccharomyces baillii, Torulaspora francisiae, Torulaspora pretoriensis, and Torulaspora globosa) are unpublished (Devin Scannell, personal communication). The gray dot in the DCR1 gene column for S. bayanus var. uvarum indicates that its DCR1 is a pseudogene.