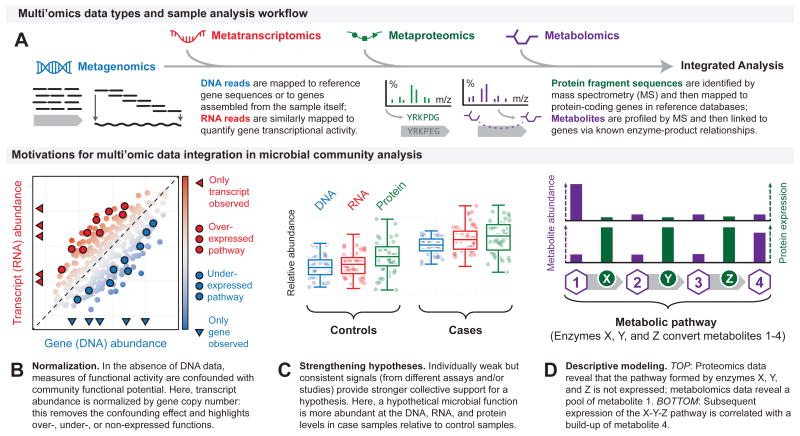
Figure 4. Integrating multi'omic data for deeper biological insights.
a. To facilitate integrated analysis of a microbiome sample, distinct multi'omic data types are often associated with microbial genes or gene families that act as a shared point-of-reference. These genes may be taken from a reference database or directly assembled from the sample. Metagenomic, metatranscriptomic, and metaproteomic sequence data (such as paired-end reads or protein fragments identified by mass spectrometry) are then directly mapped to these genes based on sequence homology, which yields information about the copy numbers and activities of genes. Metabolites (identified by mass spectrometry) can be mapped to a subset of the genes by taking advantage of known relationships between enzyme-coding genes and their products, thus providing an additional, independent measure of gene activity. There are several motivations and advantages to perform multi'omic data integration. For example, in the absence of DNA data, measures of functional activity are confounded with community functional potential. Therefore, transcript abundance can be normalized by gene copy number; this removes the confounding effect and highlights over-, under-, or non-expressed functions (part b). Individually weak but consistent signals (from different assays and/or studies) provide stronger collective support for a hypothesis. Here, a hypothetical microbial function is more abundant at the DNA, RNA, and protein levels in case samples relative to control samples (part c). Data integration also enables descriptive modeling. For example, combining data from proteomics and metabolomics analyses can reveal whether a pathway formed by different enzymes (in this case X, Y, and Z, which metabolize substrates 1, 2 and 3, respectively) is inactive or active (part d).