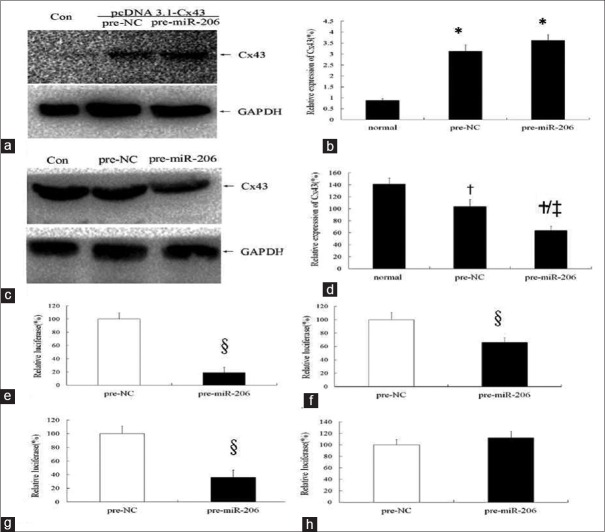
Figure 4.
The possible binding sites of connexin 43 for microRNA-206. (a-b) MicroRNA-206 showed no regulatory effects on connexin 43 of 293 cells lack of 3’UTR. a: Western blotting image; b: Statistic graph of graph a. *P < 0.05, compared with the normal group; pre-NC: Negative control group. Con: Normal group; Pre-NC: Negative control group; Pre-microRNA-206: MicroRNA-206 transfected group. c: Western blotting image; d: Statistic graph of graph c. †P < 0.05, compared with normal group; ‡P < 0.05, compared with negative control group; Con: normal group; Pre-NC: Negative control group; Pre-microRNA-206: MicroRNA-206 transfected group. (e-h) Results of luciferase activity assay. e: wild type of Cx43-3’UTR; f: mutated at the first binding sites in Cx43-3’UTR (position 480-483); g: mutated at the second binding sites in Cx43-3’UTR (position 1611-1614); h: mutated at the first and second binding sites in Cx43-3’UTR (position 480-483 and 1611-1614); pre-NC: negative control group; pre-miR-206: transfected by miR-206 group. 3’UTR: 3’ untranslated region. §ìP < 0.05, compared with pre-NC group. For the wild-type Cx43-3’UTR luciferase reporter plasmid, miR-206 can inhibit the luciferase expression (e, P <0.05). With position 480-483 mutation or 1611-1613 mutation, luciferase expression increased to 65.89% or 35.81%, respectively (f-g, P <0.05). The inhibition disappeared with mutations at positions 480-483 and 1611-1613 (h, P > 0.05).