FIG 1.
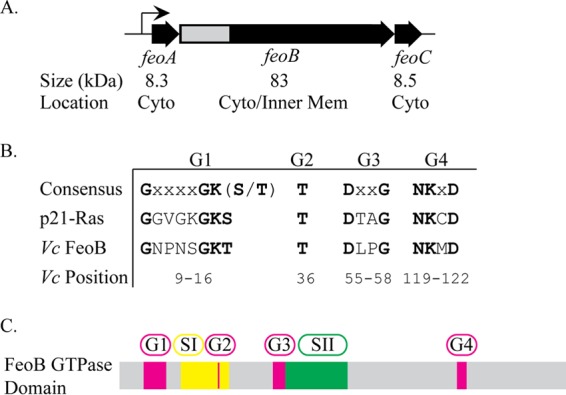
Genetic organization of V. cholerae feoABC and conserved residues of the FeoB GTPase. (A) Genetic map of V. cholerae feoABC. Below each gene are listed the predicted protein sizes and the subcellular location in the either cytoplasm (Cyto) or inner membrane (Inner Mem). The gray region indicates the section of feoB encoding the GTPase domain that is shown in more detail in panel C. (B) Consensus sequences of conserved GTPase sequence elements compared to human p21-Ras and V. cholerae FeoB, with conserved residues shown in bold. The positions of amino acid residues of each motif in V. cholerae FeoB are indicated. No G5 motif sequence has been identified in FeoB through sequence alignment. (C) Organization of the conserved GTPase sequence elements and switch regions, switch I and switch II, within the FeoB GTPase domain. The locations of switch regions I (amino acids 24 to 39) and II (amino acids 59 to 81) were predicted by sequence alignment.
