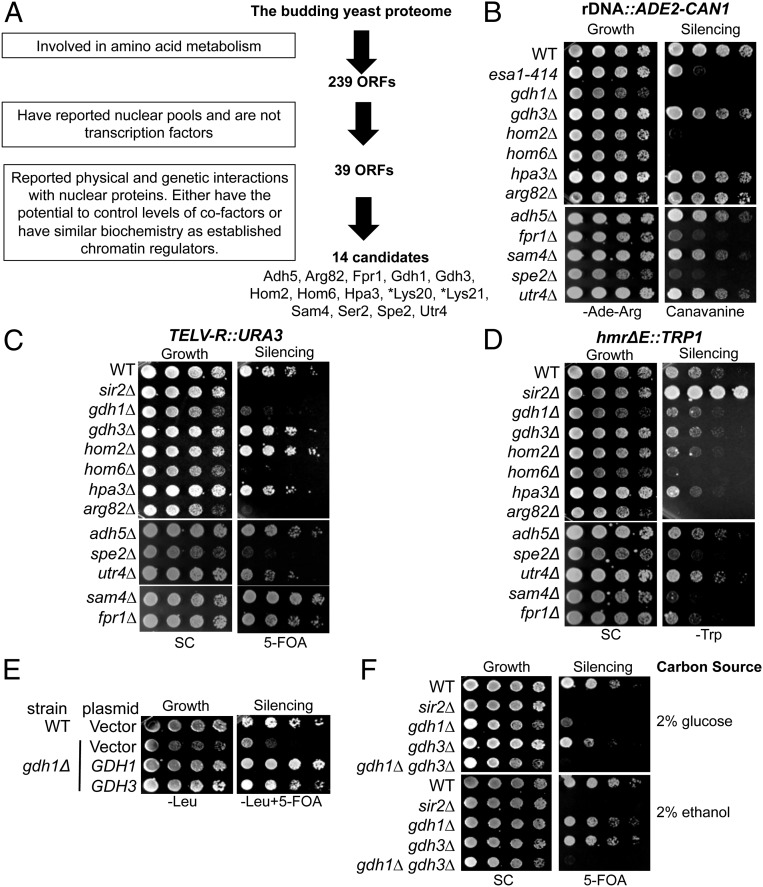
Fig. 1.
A number of amino acid metabolic proteins regulate chromatin silencing. (A) The in silico screen identified 14 proteins that potentially function in chromatin regulation. The search was performed using the Advanced Search Engine in the Saccharomyces Genome Database: yeastmine.yeastgenome.org/yeastmine/begin.do. Search criteria for each step were boxed at Left. (B–D) Strains carrying triple silencing reporters (Table S3) were plated on the indicated media to assess chromatin silencing. The esa1-414 and sir2Δ strains served as controls with established silencing defects. (B) Deletion of FPR1, GDH1, HOM2, HOM6, and SPE2 caused defects in rDNA reporter silencing. Decreased growth on canavanine indicates defective rDNA silencing. (C) Deletion of ARG82, GDH1, HOM6, and SPE2 caused defective telomeric silencing, whereas deletion of SAM4 improved silencing. Reduced growth on 5-FOA indicates defective telomeric silencing. (D) Deletion of ARG82, HOM6, SAM4, and SPE2 enhanced HMR silencing. Reduced growth on SC-Trp indicates enhanced HMR silencing. (E) Increased dosage of GDH3 suppressed gdh1Δ’s silencing defect. WT and gdh1Δ strains were transformed with 2-μ plasmids: vector (pRS425), GDH1 (pLP2764), or GDH3 (pLP2662). (F) GDH1 and GDH3 single and double mutants showed different telomeric silencing phenotypes on glucose and ethanol-containing media. WT (LPY4916), sir2∆ (LPY4979), gdh1∆ (LPY16033), gdh3∆ (LPY16785), and gdh1∆ gdh3∆ (LPY17916) strains were plated on 5-FOA with the indicated carbon sources. *Lys20 and Lys21 were identified in the screen but are not analyzed further here because of their previous characterization (14).