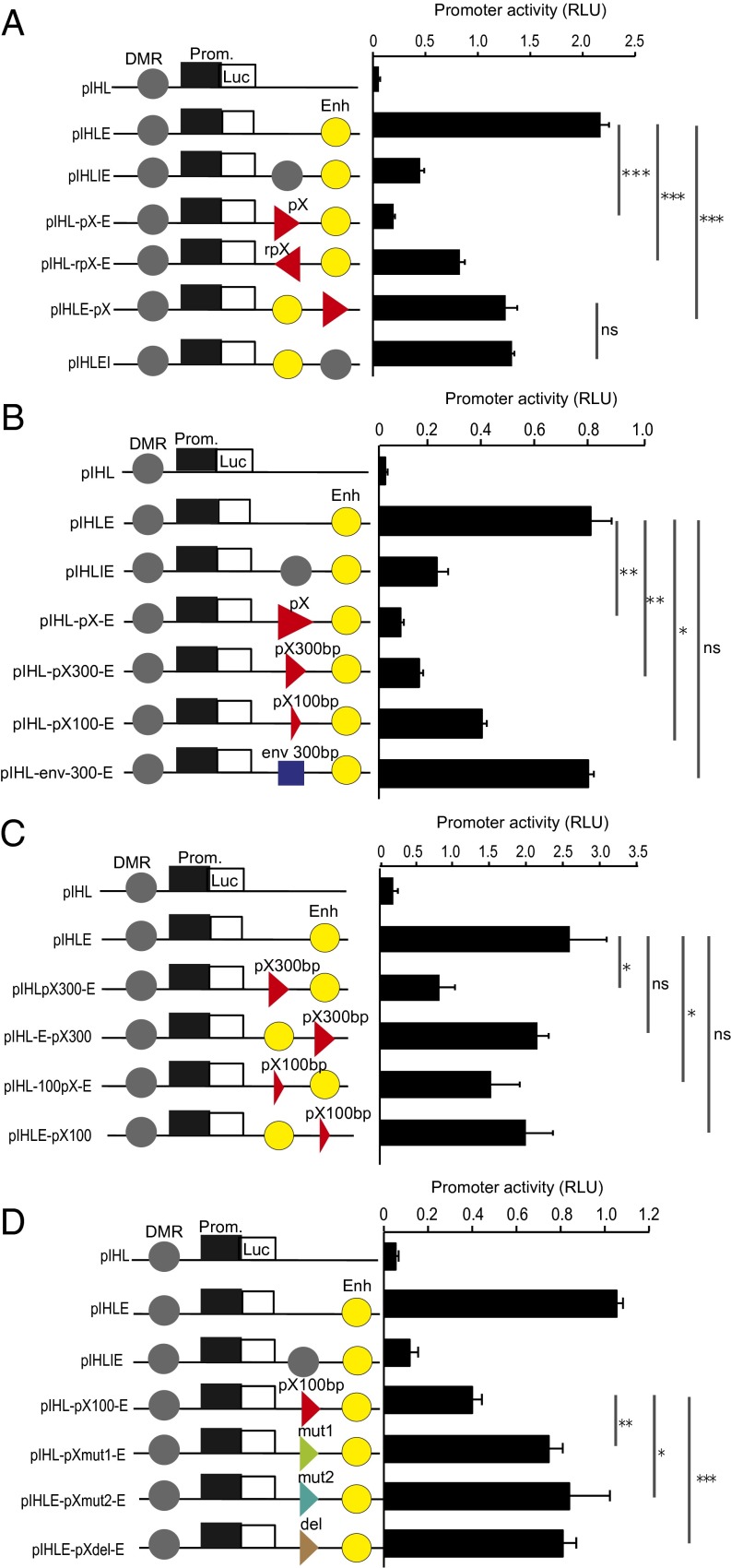
Fig. 4.
HTLV-1 pX region exerts CTCF-dependent EB activity. (A) pIHL-pX-E and pIHL-rpX-E plasmids were constructed by inserting a fragment of ∼1,000 bp, containing the CTCF-BS from the pX region, inserted in either a sense or antisense orientation between the promoter and the enhancer in pIHLE reporter vector. The H19 DMR insulator was used as a positive control for insulator function. For pIHLE-pX, the pX fragment was inserted downstream of the enhancer in pIHLE reporter vector. Firefly luciferase activities were normalized to Renilla luciferase and are shown as relative light units (RLU). (B) Plasmids pIHL-pX300-E and pIHL-pX100-E were constructed by inserting fragments, containing the pX CTCF-BS, of ∼300 bp and ∼100 bp, respectively. The pIHL-env-300-E was constructed by inserting a 300-bp fragment from the HTLV-1 env region. (C) For pIHLE-pX300 and pIHLE-pX100, the pX fragment was inserted downstream of the enhancer. Plasmids linearized with the restriction enzyme MluI were used in the assay. (D) Plasmids pIHLpXmut1-E and pIHLpXmut2-E were generated by inserting a DNA fragment of 100 bp containing the CTCF-BS with the same nucleotide substitutions as shown in Fig. S6B. In pIHL-pXdel-E, the core CTCF-BS was deleted. DMR, H19 DMR insulator; Enh, SV40 enhancer; Luc, luciferase gene; Prom., promoter of H19 gene. Data shown are representative of three independent experiments (t test: *P < 0.05; **P < 0.01; ***P < 0.001). ns, not significant.