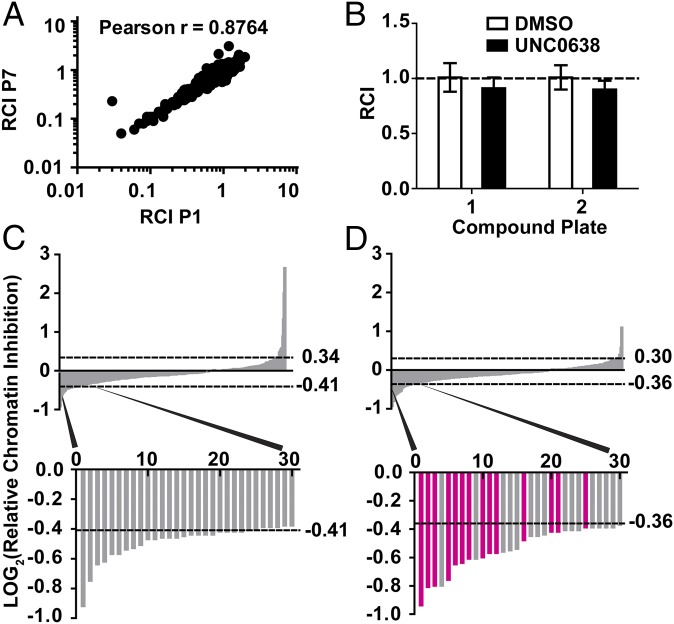
Fig. 2.
Chromatin signature-based screen identified a cluster of HDAC inhibitors that significantly decreased EWSR1-FLI1–dependent chromatin accessibility. (A) The LOG10-transformed relative chromatin inhibition (RCI) scores for the Ewing sarcoma-specific P1 and P7 regions are plotted (Pearson r = 0.8764). (B) RCI scores for DMSO and a negative control compound, UNC0638, are plotted for each 384-well plate. An RCI score of 1.0 indicates no change (dotted line). Error bars are the SD of 16 replicates. (C and D) Plate 1 (C) or plate 2 (D) LOG2 ratio of the RCI values was plotted against the rank order of compounds from greatest relative decrease (Top, left side of x axis) to the greatest relative increase (Top, right side of x axis) in FAIRE signal. The dashed lines indicate the significance cutoff of RCI values ≥2 SDs or ≤2 SDs from the average RCI for DMSO controls. Thirty compounds that show the greatest decrease in FAIRE signal for each plate are magnified (Bottom graph). The bars representing HDAC inhibitors are highlighted in magenta.