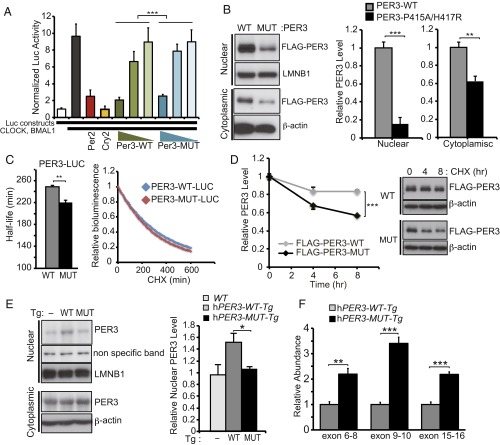
Fig. 3.
P415A/H417R variants destabilize PER3. (A) Repressor activity of WT or P415A/H417R PER3 protein on CLOCK-BMAL1–mediated expression of Per2 promoter-driven luciferase was examined by a reporter assay performed in HEK293T cells (n = 3). Luciferase activity was normalized to activity levels in the absence of CLOCK and BMAL1 (white column). Addition of CLOCK and BMAL1 resulted in a nearly 10-fold increase in luciferase activity (black column). PER3-P415A/H417R had reduced repressor activity across different titers compared with PER3-WT. PER2 (red column) and CRY2 (yellow column) were positive controls for repressor activity. ***P < 0.001, two-way ANOVA. (B) PER3 protein levels in the nuclear and the cytoplasmic fractions. HEK293T cells were transfected with indicated plasmid vectors. At 48 h after transfection, the cells were harvested and then fractionated into nuclear and cytoplasmic fractions. Results are expressed as mean ± SEM (n = 3). **P < 0.01, ***P < 0.005, Student′s t test. (C) HEK293T cells were transfected with indicated plasmid vectors. At 24 h after transfection, the culture medium was replaced by measuring medium containing CHX (100 µg/mL). Bioluminescence was measured at 10-min intervals. Bioluminescence at time point 0 was set to 1. Half-lives of PER3-LUCs were calculated by fitting the bioluminescence signals to exponential functions. Error bars show mean ± SEM (n = 3). **P < 0.01, Student’s t test. (D) HEK293T cells were transfected with indicated plasmid vectors. At 40 h after transfection, the cells were treated with CHX (100 µg/mL) for 4 or 8 h. PER3 protein levels at time point 0 were set to 1, and relative PER3 protein levels were plotted. Error bars show mean ± SEM (n = 3). ***P < 0.005, Student’s t test. (E) Western blots of nuclear and cytoplasmic fractions of liver extracts from hPER3-WT-Tg and hPER3-P415A/H417R-Tg mice at ZT12. LMNB1 and β-actin served as loading controls in the nuclear and cytoplasmic fractions, respectively. Nuclear PER3 protein levels were normalized to LMNB1 levels and are expressed as mean ± SEM (n = 3 per genotype). *P < 0.05, Student′s t test. (F) Expression levels of hPER3 mRNA in liver extracts from hPER3-WT-Tg and hPER3-P415A/H417R-Tg animals (n = 5 per genotype), as assayed by three independent TaqMan qPCR assays across different exon boundaries. Expression levels in hPER3-P415A/H417R-Tg mice were normalized to those in hPER3-WT-Tg mice. Results are expressed as mean ± SEM. **P < 0.01, ***P < 0.001, Student’s t test.