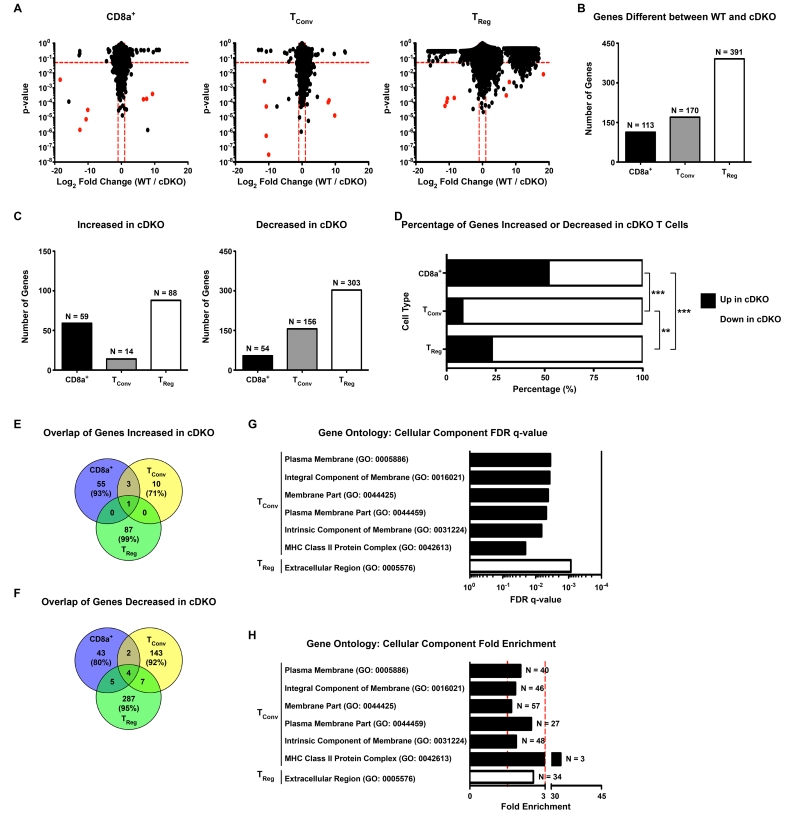
Figure 3.
Snai2 and Snai3 regulate unique gene programs among T cell lineages. (A) Graphical depiction of RNA-seq data from WT and cDKO (Left) CD8α+, (Middle) TConv and (Right) TReg cells. The y-axis represents p-value and the x-axis shows log2 fold chance (WT / cDKO). Dotted red lines mark cut-offs for significantly altered genes (p-value ≤ 0.05, WT / cDKO log2 fold change ≥ |1|). Red dots indicate Y-linked and X-chromosome inactivation-associated genes as WT donors were female and cDKO donors were male. (B) Quantification of the number of significantly altered genes upon comparison of transcriptomes from WT and cDKO CD8α+, TConv and TReg cells. Bars represent the number of genes with the exact number (N=) indicated above each bar. (C) The number of genes significantly (Left) increased or (Right) decreased in cDKO CD8α+, TConv and TReg cells. Bars represent the number of genes with the exact number (N=) indicated above each bar. (D) Percentage of genes significantly increased or decreased in cDKO CD8α+, TConv and TReg cells. Black bars represent the percentage of genes increased while white bars indicate the percentage of genes decreased. (E,F) Venn diagrams depicting the overlap of genes significantly (E) increased or (F) decreased in cDKO CD8α+, TConv and TReg cells. Number of genes and percentage of total genes (in parentheses) are indicated for each cell type. (G, H) Gene ontology analysis of cDKO dysregulated genes showing enrichment for various Cellular Component categories represented within TConv and TReg significantly altered gene sets. (G) Bars represent FDR q-values for each associated category. (H) Bars represent fold enrichment for each associated category. Vertical dotted red lines demarcate 1.5- and 3-fold enrichment, respectively. The number of contributing number of genes (N=) for each category is indicated next to each bar. (A-H) Number of animals: N = 4 for all cell types except for cDKO TRegs (N = 3). (D) Fisher’s Exact Test: ** p ≤ 0.01, *** p ≤ 0.001.