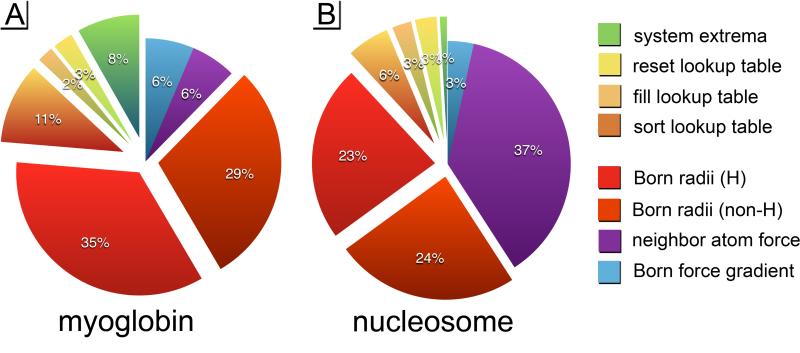
Figure 3.
These are the approximate distributions of GPU time spent on two systems when run the CUDA-GBSW algorithm: A) myoglobin with 2459 atoms, and B) the eukaryotic nucleosome with 22481 atoms. Notice that the neighbor lookup table is the only kernel that doesn't scale approximately with O(N) complexity. In larger systems the neighbor-atom force becomes the most expensive part of the solvent model calculation.