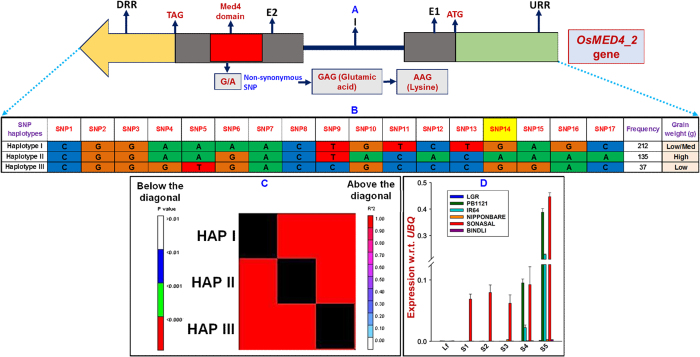
Figure 5. The molecular haplotyping and SNP haplotype-specific association analysis/LD mapping in an OsMED4_2 gene (A) validating its strong association potential for grain weight/size differentiation in rice. The genotyping of 17 SNPs, including one missense non-synonymous SNP (G/A, shaded with yellow colour) [encoding for Glutamic acid (GAG) to Lysine (AAG)] among 384 rice accessions (association panel) constituted three haplotypes (B). (C) Three SNP haplotype marker-based genotyping information produced a higher LD estimate and resolution covering the entire gene. (D) The differential expression profiling of OsMED4_2 gene in five seed developmental stages (S1–S5) and flag leaves (Lf) of six contrasting low (Sonasal and Bindli) and high (LGR, PusaBasmati 1121, Nipponbare and IR 64) grain weight rice accessions.
E1: Exon1, E2: Exon2, I: Intron, URR: upstream regulatory region and DRR: downstream regulatory region.