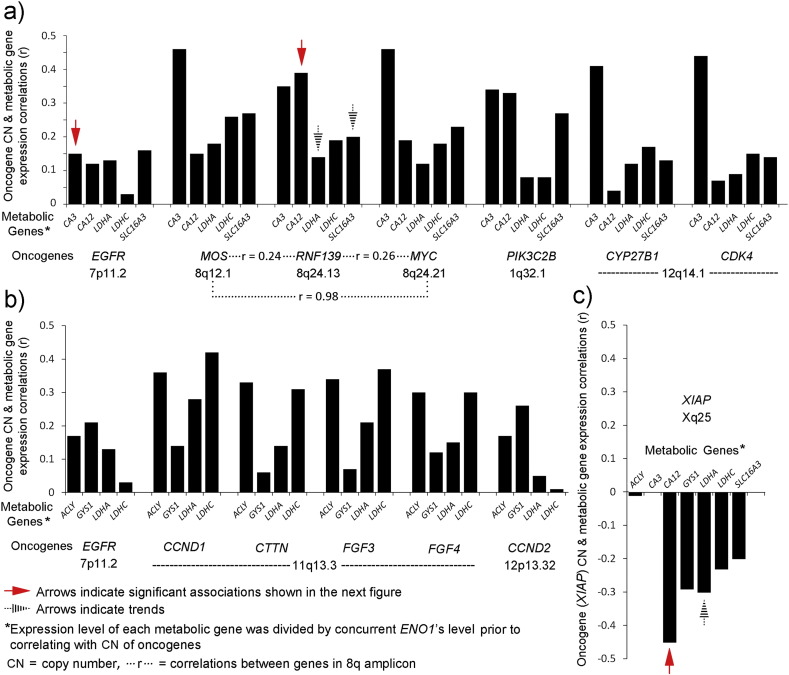
Fig. 6.
Patterns of functional relevance in the correlations between metabolic genes and oncogenes. (a) A pattern attributed to genes whose expressions protect glycolysis in acidic conditions, possibly to supra-physiologic limits. Positive correlations of the ENO1 transformed expressions of metabolic genes (CA3, CA12, LDHA, LDHC, and SLC16A3(or MCT4)) occurred with CN of oncogenes (EGFR, MOS, RNF139, MYC, PIK3C2B, CYP27B1, and CDK4). Those supported by significant associations (Wilcoxon Rank Sum, Section 3.6) are indicated by solid red arrows and trends by black striped arrows. Complexity within a potential 8q amplicon is indicated by strong correlation (r = 0.98) between MOS and MYC but weaker correlations between each of them and intervening RNF139. (b) A pattern attributed to non-aerobic glycolysis operating in intermittent avascular conditions based on previous studies of glycolytic-dependent glioblastoma cell migration [9], [11], involving ENO1 transformed expressions of metabolic genes, ACLY, GYS1, LDHA, and LDHA, was identified by shared positive correlations with CN of oncogenes, EGFR, CCND1, CTTN, FGF3, FGF4, and CCND2. (c) The CN gain of XIAP revealed an opposing pattern for metabolic genes' expressions. The significant association (Wilcoxon Rank Sum, Section 3.6) and a trend are indicated by the solid red and striped black arrows, respectively.