Figure 1.
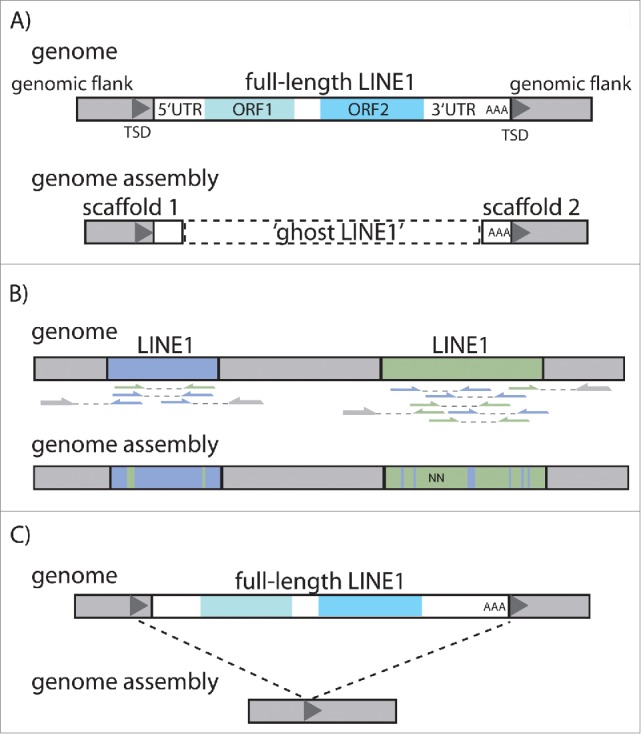
Selected problems that can affect TE sequences and copy numbers in a final genome assembly. (A) Due to different assembly algorithms implemented in the programs, problematic regions might be left out of assemblies all together due to the lack of supporting or conflicting reads leading to ghost LINE1s. In such cases, full-length LINE1s are split across 2 different scaffolds. (B) Paired-end reads (opposing arrows connected by dashed lines) originating from multiple LINE1 copies can falsely be incorporated into one single consensus sequence, leading to unintentional substitutions as well as ambiguous nucleotides (N) in the TE sequence in the assembly. (C) The identical flanking target site duplications (TSDs) can be falsely assembled into a single sequence, collapsing the entire LINE1 (or ERV). TSD: target site duplication; AAA: poly(A)-tail; ORF: open reading frame, UTR: untranslated region.
