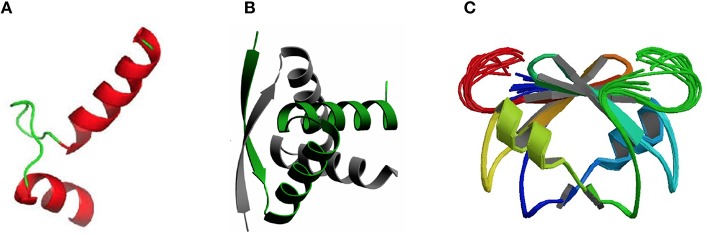
Figure 1.
Three-dimensional structures showing the most frequent DNA-binding domains found in prokaryotic type II antitoxin proteins. The most frequent DNA-binding domains found in type II antitoxins include: (A) the HTH-motif (of which the smallest structural motif is shown) that has two α-helices (red) connected by a small loop (green); (B) the RHH folding motif, in which the minimal structure (as the one depicted corresponding to the CopG transcriptional repressor) is generated by two antiparallel β-strands (arrows, with arrowheads pointing to the C-terminal part of the protomer) that generate a ribbon; each strand comes from one of two protein monomers and they are involved both in dimer formation and in specific interactions with the DNA bases in the antitoxin DNA target (adapted from Gomis-Rüth et al., 1998), and (C) the SpoVT/Abr DNA binding motif in which the dimeric molecules are constructed by three- and four-stranded antiparallel β-sheets (upper part of the molecule) that are tightly packed, generating the DNA-binding domain. Loops keeping the outer part of the molecules are indicated by light green and red colors.