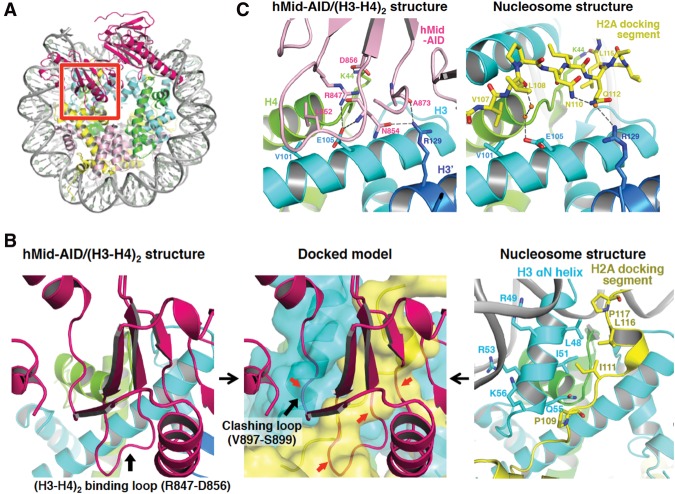
Figure 4.
Structural model of H2A–H2B displacement from nucleosomes by hMid–AID. (A) Docking model constructed by making the best superposition of the (H3–H4)2 structure between hMid–AID/(H3–H4)2 and the nucleosome (PDB code 2CV5). hMid is colored deep pink. The red box represents a close-up view shown in the middle panel of B. (B) H2A–H2B is displaced from nucleosomes by steric hindrances. (Left panel) The (H3–H4)2 binding loop on hMid–AID (deep pink) interacts with (H3–H4)2 in the hMid–AID/(H3–H4)2 structure (black arrow). (Middle panel) The clashing loop on hMid–AID collides with the H3 αN helix in the docking model (black arrow). (Middle and right panels) Both the (H3–H4)2-binding loop and the clashing loop on hMid–AID cause steric hindrances (red arrows in the middle panel) around the H2A-docking segment (yellow) and the H3 αN helix (cyan), which form hydrophobic interactions to tether H2A–H2B to (H3–H4)2 in the nucleosome (right panel). (C) Detailed views of hMid–AID in the hMid–AID/(H3–H4)2 complex (left panel) and the H2A-docking segment in nucleosomes (right panel) on the contact surface of (H3–H4)2 (colored as in Fig. 2A). They share interactions with key residues (Val101 and Glu105 of H3, Arg129 of H3′, and Lys44 of H4). Hydrogen bonds are indicated by gray dashed lines, and contact residues are depicted. Water molecules are shown as orange balls.