Abstract
Toxoplasma gondii remains a global public health problem. However, its pathophysiology is still not-completely understood particularly the impact of infection on host liver metabolism. We performed iTRAQ-based proteomic analysis to evaluate early liver protein responses in BALB/c mice following infection with T. gondii PYS strain (genotype ToxoDB#9) infection. Our data revealed modification of protein expression in key metabolic pathways, as indicated by the upregulation of immune response and downregulation of mitochondrial respiratory chain, and the metabolism of fatty acids, lipids and xenobiotics. T. gondii seems to hijack host PPAR signaling pathway to downregulate the metabolism of fatty acids, lipids and energy in the liver. The metabolism of over 400 substances was affected by the downregulation of genes involved in xenobiotic metabolism. The top 10 transcription factors used by upregulated genes were Stat2, Stat1, Irf2, Irf1, Sp2, Egr1, Stat3, Klf4, Elf1 and Gabpa, while the top 10 transcription factors of downregulated genes were Hnf4A, Ewsr1, Fli1, Hnf4g, Nr2f1, Pparg, Rxra, Hnf1A, Foxa1 and Foxo1. These findings indicate global reprogramming of the metabolism of the mouse liver after acute T. gondii infection. Functional characterization of the altered proteins may enhance understanding of the host responses to T. gondii infection and lead to the identification of new therapeutic targets.
Introduction
Over one-third of human population worldwide are chronically infected with the apicomplexan protozoan parasite Toxoplasma gondii [1], and are thus at high risk of developing serious diseases if they became immune-compromised or when women get pregnant. Despite great effort and much progress, toxoplasmosis remains a major threat to global health. No vaccine is available to date; anti-toxoplasma drugs are not highly effective and are associated with significant side effects [2]. Also, the parasite is capable of developing resistance to these drugs. Evidently, the current toxoplasmosis treatments have limitations. Novel approaches and new therapeutic targets need to be explored. The wide range of susceptible hosts and the remarkable ability of T. gondii to infect and replicate within almost any nucleated cell [1], resulted in T. gondii being associated with a variety of clinical disorders. However, brain and spinal cord have been always the primary targets of T. gondii infection.
Interestingly, many studies have also indicated that the liver is another target organ for T. gondii replication, as evidenced by the several hepatic pathologies, such as hepatomegaly, hepatitis, granuloma [3], necrosis, cholestatic jaundice [4], and cirrhosis [5,6], associated with T. gondii infection. Also, T. gondii infection has been incriminated in causing abnormal liver function tests in mice, including alanine aminotransferase (ALT) and aspartate aminotransferase (AST) six days post-infection in mice [7] and liver dysfunction in liver and kidney of transplant recipients [8]. Acute T. gondii infection in mice with RH strain revealed an association between the increased number of hepatic stellate cells (HSCs) and the amount of T. gondii antigens, suggesting a modulatory role for HSCs in the pathogenesis of T. gondii-induced hepatopathy [9]. Furthermore, T. gondii infection can induce pharmacokinetic changes [10,11]. These data indicate that hepatic toxoplasmosis does exist, but probably because T. gondii spreads to the liver during the acute phase of infection it may go unnoticed.
Although it is generally known that parasitic and host factors contribute to the pathogenesis and progression of toxoplasmosis, the interplay between T. gondii and host liver remains poorly understood. This is surprising giving the fact that liver is an essential organ that can carry out a wide variety of critical operations, such as detoxification of xenobiotics and producing bile that facilitates digestion. Functional change in the liver could result in severe disease, such as cancer, malnutrition, hepatitis, jaundice, cirrhosis, and adverse drug reactions [12]. Different tools have been used to investigate molecular mechanisms of toxoplasmosis, including proteomics [13–15], transcriptomics [16,17], and metabolomics [18,19]. Specifically, proteomics can provide high-throughput methods such that hundreds of proteins can be identified and/or quantified in a single analysis.
Mass spectrometry-based proteomic approaches have significantly matured in the last few years and have greatly facilitated investigations of host proteome during T. gondii infection. For example, proteome of T. gondii-infected macrophage from Kunming mice 24 hour post infection exhibited altered expression of 38 proteins involved in metabolism, cell structure, signal transduction and immune response [14]. Similar proteomic profile was obtained from brain tissues of Kunming mice infected with genotype II T. gondii PRU strain [15]. Also, proteome of human foreskin fibroblasts (HFFs) infected with genotype II T. gondii RH strain revealed differentially expressed proteins involved in many metabolic processes including glycolysis, lipid metabolism, cell death, stress, and immune response [13]. As can be gleamed from the wealth of proteomics-based information available, this approach has contributed greatly to our understanding of molecular mechanisms underlying host response to T. gondii infection and clearly indicates global reprogramming of host cell metabolism by the parasite. However, previous studies have been based on two-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis [13–15], which is relatively low throughput for protein identification or quantification. Also, there are no papers reporting the use of iTRAQ-based proteomics to assay mouse liver samples for hepatic toxoplasmosis investigation. Animal models have been extremely helpful for providing fundamental insights into the pathogenesis of T. gondii infection. While some of the features of specific models may not fully mimic human pathophysiological changes, animal models provide a system to understand some of the basic host responses to infection. The aim of the present study was to investigate the global proteomic changes in the liver of mice infected with T. gondii. The unbiased iTRAQ proteomic method was employed to understand mechanisms of liver injury that occur during acute infection with T. gondii.
Materials and Methods
Ethical statement
All animal procedures were approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Permit No. LVRIAEC2013-006). All experiments were performed in strict compliance with requirements of the Animal Ethics Procedures and Guidelines of the People's Republic of China.
Mice, parasite challenge and sample collection
The PYS strain of T. gondii (genotype ToxoDB#9) the most widespread genotype in China [13], was isolated and maintained in our laboratory. BALB/c mice were purchased from the Laboratory Animal Center of Lanzhou Veterinary Research Institute, Chinese Academy of Agriculture Science. The health status and behavior of mice were examined daily. Mice had ad libitum access to food and water. Six-week-old female BALB/c mice were randomly divided into two equal groups (3 mice each group). Each mouse of the infected group was injected intraperitoneally (i.p.) with 200 tachyzoites of the PYS strain counted by hemocytometer, whereas mice in the control group were mock-inoculated with phosphate buffer saline (PBS, pH7.4) without tachyzoites. Six days post-infection (PI) mice were humanely sacrificed by CO2 asphyxiation. The liver tissues were collected and stored at -80°C until use. T. gondii infection was confirmed by microscopic examination and PCR of liver DNA as described previously [20]. PCR positive products were submitted to Genewiz Company (Beijing, China) for sequencing using ABI3730 sequencer on both strands.
Protein extraction
Total protein from the each cryo-preserved mouse liver samples was prepared as follows. Each biological sample was individually ground to powder in liquid nitrogen and extracted in Lysis buffer 1 (7 M Urea, 2 M Thiourea, 4% CHAPS, 40 mM Tris-HCl, pH 8.5) containing 1 mM PMSF and 2 mM EDTA (final concentration). After 5 min, 10 mM DTT (final concentration) was added to the sample. The suspension was sonicated at 200 W for 15 min and then centrifuged at 4°C, 30,000 g for 15 min. The supernatant was mixed with 5 volumes of chilled acetone containing 10% (v/v) TCA and incubated at -20°C overnight. After centrifugation at 4°C, 30,000 g for 15 min, the supernatant was discarded. Then, the precipitate was washed with chilled acetone three times. The pellet was air-dried and dissolved in Lysis buffer 2 (7 M urea, 2 M thiourea, 4% NP40, 20 mM Tris-HCl, pH 8.0–8.5). The suspension was sonicated at 200 W for 15 min and centrifuged at 4°C, 30,000 g for 15 min. The supernatant was then transferred to another tube. To reduce disulfide bonds in proteins of the supernatant, 10 mM DTT (final concentration) was added and incubated at 56°C for 1 hour. Subsequently, 55 mM IAM (final concentration) was added followed by incubation for 1 hour in the dark. The supernatant was mixed with 55 volumes of chilled acetone for 2 hour at -20°C to precipitate proteins. After centrifugation at 4°C, 30,000 g for 15 min, the supernatant was discarded and the pellet was air-dried for 5 min, dissolved in 500 μl 0.5 M TEAB (Applied Biosystems, Milan, Italy), and sonicated at 200 W for 15 min. Finally, samples were centrifuged at 4°C, 30,000 g for 15 min. The supernatant was transferred to a new tube and proteins were quantified, using the Bradford method [21]. The proteins in the supernatant were stored at -80°C until further analysis.
iTRAQ labeling and SCX fractionation
Proteins (100 μg) from each sample were digested with Trypsin Gold (Promega, Madison, WI, USA) with the ratio of protein:trypsin = 30:1 at 37°C for 16 hour. After trypsin digestion, peptides were dried by vacuum centrifugation and were reconstituted in 0.5 M TEAB and processed according to the manufacture’s protocol for 8-plex iTRAQ reagent (Applied Biosystems).
SCX chromatography was performed with an LC-20AB HPLC Pump system (Shimadzu, Kyoto, Japan). The iTRAQ-labeled peptide mixtures were reconstituted with 4 ml buffer A (25 mM NaH2PO4 in 25% ACN, pH 2.7) and loaded onto a 4.6×250 mm Ultremex SCX column containing 5 μm particles (Phenomenex). The peptides were eluted at a flow rate of 1 ml/min with a gradient of buffer A for 10 min, 5–60% buffer B (25 mM NaH2PO4, 1 M KCl in 25% ACN, pH 2.7) for 27 min, 60–100% buffer B for 1 min. The system was then maintained at 100% buffer B for 1 min before equilibrating with buffer A for 10 min prior to the next injection. Elution was monitored by measuring the absorbance at 214 nm, and fractions were collected every 1 min. The eluted peptides were pooled into 20 fractions, desalted with a Strata X C18 column (Phenomenex) and vacuum-dried.
LC-ESI-MS/MS analysis based on Triple TOF 5600
Each fraction was resuspended in buffer A (5% ACN, 0.1% FA) and centrifuged at 20,000 g for 10 min, and the final concentration of peptide was 0.5 μg/μl. About 10 μl supernatant was loaded on a LC-20AD nanoHPLC (Shimadzu, Kyoto, Japan) by the auto sampler onto a 2 cm C18 trap column. Then, the peptides were eluted onto a 10 cm analytical C18 column (inner diameter 75 μm) packed in-house. The samples were loaded at 8 μL/min for 4 min, then the 35 min gradient was run at 300 nl/min starting from 2 and increasing to 35% buffer B (95% ACN, 0.1% FA), followed by 5 min linear gradient to 60%, followed by 2min linear gradient to 80%, and maintenance at 80% buffer B for 4 min, and finally a return to 5% for 1 min.
Data acquisition was performed with a TripleTOF 5600 System (AB SCIEX, Concord, ON) fitted with a Nanospray III source (AB SCIEX, Concord, ON) and a pulled quartz tip as the emitter (New Objectives, Woburn, MA). Data was acquired using an ion spray voltage of 2.5 kV, curtain gas of 30 psi, nebulizer gas of 15 psi, and an interface heater temperature of 150°C. The MS was operated with a RP of greater than or equal to 30,000 FWHM for TOF MS scans. For IDA, survey scans were acquired in 250 ms and as many as 30 product ion scans were collected if exceeding a threshold of 120 counts per second (counts/s) and with a 2+ to 5+ charge-state. Total cycle time was fixed to 3.3 s. Q2 transmission window was 100 Da for 100%. Four time bins were summed for each scan at a pulser frequency value of 11 kHz through monitoring of the 40 GHz multichannel TDC detector with four-anode channel detect ion. A sweeping collision energy setting of 35±5 eV coupled with iTRAQ adjust rolling collision energy was applied to all precursor ions for collision-induced dissociation. Dynamic exclusion was set for 1/2 of peak width (15 s), and then the precursor was refreshed off the exclusion list.
Proteomic data analysis
Discoverer 1.2 (PD 1.2, Thermo), 5600 msconverter and the MGF file were searched. Protein identification was performed by using the Mascot search engine (Matrix Science, London, UK; version 2.3.02) against the Uniprot database. For protein identification and quantitation, a protein was required to contain at least two unique peptides. The peptide for quantification was automatically selected by the algorithm to calculate the reporter peak area, error factor (EF) and p-value (default parameters in Mascot Software package). Student’s t-test was performed using the Mascot software. The resulting data set was auto bias-corrected to the biological replicate. The quantitative protein ratios were weighted and normalized by the median ratio in Mascot. We only used ratios with p values < 0.05, and only fold change >1.5 was considered as significant.
Functional enrichment of the differentially expressed proteins (DEPs) or cellular organelle components was conducted using a cytoscape plugin, BiNGO [22], and all data files used in BiNGO were downloaded from the Gene Ontology database (GO, http://www.geneontology.org/) [23]. Pathway analyses of DEPs were performed through the KEGG database (KEGG, http://www.genome.jp/kegg/) [24]. Hypergeometric test and the Benjamini-Hocherg False Discovery Rate (FDR) correction were used for identifying significantly enriched GO terms or KEGG pathways. Significance levels of p < 0.05, following FDR correction, were used as cut-offs for the enrichment threshold of GO term or KEGG pathway. Differently expressed GO terms or KEGG pathways were also identified where the FDR corrected p value was less than 0.05, the number of DEPs was greater than 3, and the ratio between upregulated protein and downregulated protein was greater than 2 or less than 0.5. The relationships among metabolism of substances, liver damage and DEPs were analyzed using the Comparative Toxicogenomics Database (http://ctdbase.org/) [25]. Transcription factor analysis of DEPs was analyzed by Pscan web service (http://159.149.160.51/pscan/) [26]. The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium [27] via the PRIDE partner repository with the dataset identifier PXD003399.
Results
Confirmation of T. gondii infection in mouse livers
All 3 biological replicates in the infected group exhibited signs indicative of toxoplasmosis, but no mouse has died of T. gondii infection. Also, T. gondii tachyzoites were observed in peritoneal fluid of all infected mice. In contrast, no clinical signs or tachyzoites of T. gondii were observed in the non-infected control mice. Liver samples from all infected mice were PCR-positive and DNA sequencing confirmed that all PCR products are amplicons of B1 gene of T. gondii. In contrast, the negative PCR control template and all DNA templates of the control mouse livers were PCR negative.
Proteomic characterization
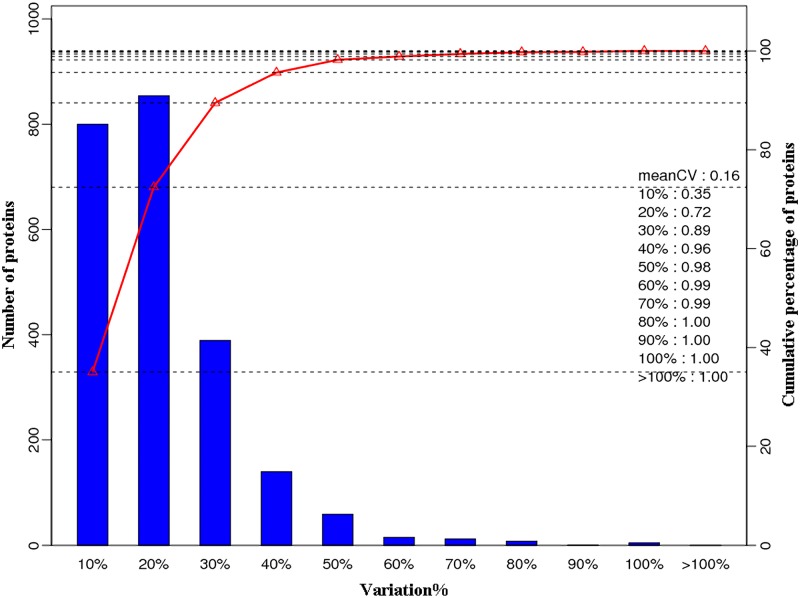
A total of 407,280 spectra were obtained. According to the Mascot program searching, 46,681 spectra were matched and 15,699 high scoring unique peptides were obtained. A total of 3,643 proteins were identified, 301 of which were differentially expressed in infected livers (p < 0.05), including 73 downregulated proteins and 228 upregulated proteins. The details of differentially expressed proteins are shown in S1 Table. The variation coefficients between three biological replicates are shown in Fig 1. Variability of 72% protein was not more than 20%.
Fig 1. Coefficients of variation values between the three biological replicates.
Bottom-axis (%variation) shows the variation of identified proteins. Left-axis shows the number of identified proteins. Right-axis and red line show the cumulative percentage of identified proteins.
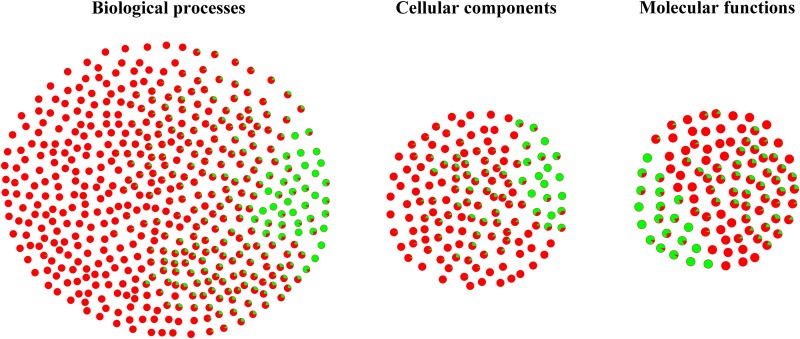
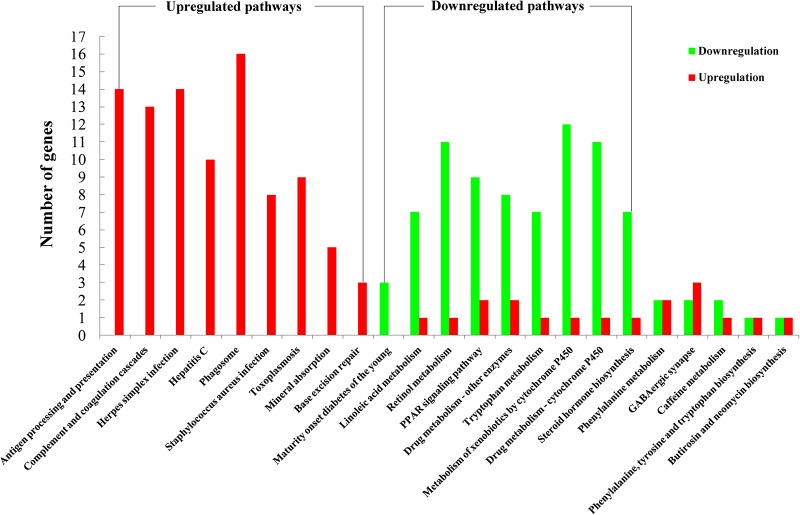
GO analysis was performed to study the biological change of infected liver. 139 cellular components, 103 molecular functions and 473 biological processes of infected liver were differentially expressed in the present study. As shown in Fig 2, most differentially expressed GO terms were upregulated, including cellular components (vesicle, nucleus, Golgi apparatus, lysosome, endosome, actin cytoskeleton) and biological processes (material transport, actin cytoskeleton organization, immune response related terms, protein complex assembly, protein folding, proteolysis, negative regulation of cell death and nucleobase-containing small molecule metabolic process). However, downregulated GO terms were also detected, such as cellular components (mitochondrial envelope, microbody), biological processes (alpha-amino acid catabolic process, oxidation-reduction process, organic acid metabolic process, fatty acid metabolic process, cellular lipid metabolic process, drug or xenobiotic metabolic process) and molecular oxidoreductase activity and heme binding. The details of differentially expressed proteins of cellular components, biological processes and molecular functions are listed in S2–S4 Tables, respectively. As shown in Fig 3, 23 pathways were significantly enriched in KEGG analysis, including 9 upregulated pathways and 9 downregulated pathways. Consistent with GO enrichment analysis, most upregulated pathways were involved in immune response, while downregulated pathways were metabolism-related.
Fig 2. Differentially expressed GO term of infected liver.
Pie dot shows the differentially expressed GO terms. Red color represents the percentage of upregulated proteins. Green color represents the percentage of downregulated proteins.
Fig 3. Enriched pathways in KEGG database.
23 pathways were enriched in the KEGG database. Red color represents upregulation. Green color represents downregulation.
Comparative toxicogenomics analysis
In the present study, we identified 15 DEPs involved in drug or xenobiotic metabolism, including Gstm1, Sult2a5, Sult2a1, Cyp2c54, Cyp3a16, Cyp3a11, Cyp8b1, Cyp2c50, Cyp2b9, Cyp4a10, Cyp3a25, Cyp1a2, Cyp2a22, Cyp2c29 and Cyp3a13. Among the DEPs, only Cyp3a13 was upregulated in mouse livers infected with T. gondii. According to the Comparative Toxicogenomics Database, over 400 substances were catabolized by the DEPs described above. The details of relationships between the DEPs and chemicals are listed in S5 Table. We also analyzed all DEPs in the Comparative Toxicogenomics Database to obtain a list of liver diseases that correlate with DEPs. Cancer, liver cirrhosis, drug-induced liver injury, cholestasis, drug eruptions, fatty liver, inflammation and necrosis were shown in the results of the Comparative Toxicogenomics Database analysis.
Transcription factor analysis of DEPs
In the present study, 2 transcription factors (Ybx1, Stat3) and 6 transcription co-factors (Calr, Ewsr1, Pml, Mybbp1a, Thrap3 and Ddx17) were upregulated. In Pscan analysis, 263 transcription factor motifs were employed to analyze the transcription factors that could be used by DEPs. The top 10 transcription factors used by the upregulated proteins were Stat2, Stat1, Irf2, Irf1, Sp2, Egr1, Stat3, Klf4, Elf1 and Gabpa, while the top 10 transcription factors used by the downregulated proteins were Hnf4A, Ewsr1, Fli1, Hnf4g, Nr2f1, Pparg, Rxra, Hnf1A, Foxa1 and Foxo1.
Discussion
Over the last decade, a significant amount of data, including serological, biochemical and direct detection studies, has suggested a potential role for T. gondii infection in liver pathologies [3–6,8–11]. However, the mechanisms involved in establishing hepatic infection with this organism remain unclear.
Unbiased iTRAQ-based proteomic analysis revealed 301 differentially expressed proteins in mouse livers and most of them were upregulated. These altered proteins play crucial roles in several cellular functions, particularly in transcription and translation processes. Our results demonstrate that host protein involved in cellular immune defense, lipid metabolism, and intracellular transport are regulated during the early stage of T. gondii infection. Potential roles of the altered proteins in response to acute T. gondii infection in the present study corroborated the current knowledge of T. gondii infection. T. gondii has developed strategies to manipulate important host cell immune pathways. Indeed, infection of mammalian cells with T. gondii induces many changes in host cell gene transcription, including those genes involved in energy metabolism, immune responses and signaling [28]. Low variability among the tested biological replicates indicates consistency in our results. GO and KEGG enrichment analyses were applied to study the biological change of mouse livers infected by T. gondii. Several GO terms were downregulated, such as the components of mitochondrial envelope, microbody, oxidation-reduction process, organic acid metabolic process, fatty acid metabolic process, cellular lipid metabolic process, drug or xenobiotic metabolic process, oxidoreductase activity and heme binding. On the other hand, the upregulation of GO terms was also detected, such as vesicle, Golgi apparatus, lysosome, endosome, material transport, actin cytoskeleton, protein complex assembly, protein folding, proteolysis, negative regulation of cell death and immune response-related terms. To the best of our knowledge, the present study is the first to deploy iTRAQ-based proteomic method to address changes in the cellular proteome of mouse liver during T. gondii infection. Our findings will be discussed in the light of how the parasite alters the immune response, as this is crucial for our understanding of how the infection is controlled, and as discussed below, the host immune response is a major target of T. gondii manipulation.
Immune response
Activation of a cell-mediated immunity plays a crucial role in protection and pathogenesis of T. gondii infection [29]. Stimulation of host cells by IFN-γ inhibits growth of T. gondii primarily by induction of indoleamine 2,3-dioxygenase (IDO) activity, which deprives the parasite of the essential amino acid tryptophan. Also, T. gondii upregulates inducible nitric oxide synthase (iNOS) to diminish arginine levels and upregulates interferon-inducible immunity related GTPases (IRGs) and guanylate binding proteins (GBPs) to disrupt the parasitophorous vacuole membrane (PVM), which separates the parasite from host immune defences [30]. IFN-γ was not detected in the present study, probably due to the limitation of the mass-spectrometric technique (i.e. at least two unique peptides are required for the identification of a specific protein and mass-spectrometry cannot identify or distinguish all proteins in a biological system [31]). However, five immunity-related GTPases (IRGs) (Gm12250, Igtp, Iigp1, Irgm1, Gm495) and five GBPs (Gbp7, Gbp6, Gbp4, Gbp2b, Gbp2) were upregulated DEPs. The roles of Irgm1, Iigp1, Igtp, Gm12250 (also known as Irgb10), Gbp7, Gbp6, Gbp2 and Gbp4 in the disruption of PVM of T. gondii have been described [32–36]. Interestingly, one indoleamine 2,3-dioxygenase (Ido2) was downregulated in the infected livers. This downregulation may have a protective role for T. gondii by preventing the decrease of host tryptophan levels. IFN-γ can be produced by IL-1β pathway activation [37]. However, in the present study, the IL-1β pathway was inhibited by the upregulation of the interleukin 1 receptor antagonist (Il1rn), which blocks the IL-1β signal [38]. Upregulation of Il1rn was not reported in previous proteomic reports [13–15]. The discrepancy between our results and the reported proteomic findings may be attributed to the use of low throughput proteomic method (e.g., DIGE) [13–15]. Regardless, the upregulation of immune response-related terms is, overall, consistent with previous proteomic reports in previous studies [13–15].
Regulatory elements and signaling pathways
Because protein expression is controlled by Cis-regulatory elements and many innate immune effectors are under the control of transcription factors in the anti-T. gondii immune response, we performed Cis-regulatory element analysis to obtain more insights into the altered biological processes of the infected liver. The top 10 transcription factors involved in regulation of upregulated genes were Stat2, Stat1, Irf2, Irf1, Sp2, Egr1, Stat3, Klf4, Elf1 and Gabpa, while the top 10 transcription factors used by downregulated genes were Hnf4A, Ewsr1, Fli1, Hnf4g, Nr2f1, Foxa1, Foxo1, Hnf1A, Pparg and Rxra.
Infection with T. gondii interferes with host cell signaling pathways implicated in protective immunity, for instance by blocking the transcription factors signal transducer and activator of transcription 1 (STAT1) [39] and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) [40]. Fibroblasts infected with any of the three clonal types of T. gondii (i.e. I, II and III) are unresponsive to the anti-parasitic effects of IFN-γ and this appears to be due to impaired STAT1 signaling [41]. In T. gondii-infected bone marrow derived macrophages, this occurs due to modifications in chromatin structure, which disrupt STAT1 binding to nuclear promoters [42]. Also, T. gondii infection upregulates anti-inflammatory pathways including those involving STAT3 [43] and the suppressor of cytokine signaling protein (SOCS) 1 [44], SOCS3 [45], potentially compromising host resistance mechanisms to T. gondii infection.
Two important transcription factors Pparg and Rxra were involved in the regulation of downregulated proteins. According to the KEGG database, Pparg (also known as Nr1c3) and Rxra (also known as Nr2b1) are critical transcription factors of the peroxisome proliferator-activated receptor (PPAR) signaling pathway, which play a key role in modulating hepatic triglyceride accumulation in liver. In our study, PPAR signaling pathway was significantly enriched (Fig 3), and 9 differentially expressed proteins, such as Ehhadh, Acsl1, Acaa1b, Cyp8b1, Me1, Apoa2, Apoc3, Cyp4a10, and Fads2 involved in PPAR signaling pathway were downregulated. These findings show that T. gondii may regulate fatty acid metabolism via regulating PPAR signaling pathway to downregulate fatty acid metabolism. Differential regulation of a variety of lipid species has also been reported previously [13].
Cell structure
The host immune responses do not operate in isolation. In fact, anti-T. gondii immune response is engaged with many cellular components and biological processes, such as cellular organelles, protein folding, protein complex assembly, proteolysis, material transport and host secretion or endocytosis. Most GO terms of these were upregulated as listed in S2–S4 Tables. Secretion, endocytosis, and phagocytosis are controlled by vesicle-mediated transport processes, which mediate exchange of materials among different cellular organelles, such as the endoplasmic reticulum (ER), Golgi apparatus, lysosome, endosome and other cellular organelles. Most importantly, this process sorts and transports proteins to where they are functionally required [46,47]. For example, MHC complexes are sorted and transported by vesicle-mediated transportation after synthesis and assembly [47]. In the present study, vesicle-mediated transport processes were upregulated. However, at mRNA level, Tanaka et al. reported that vesicle-mediated transport process was downregulated in mouse brains infected by T. godnii genotype I strain [48]. This inconsistency may be related to the use of different host tissues, different T. gondii strains or experimental conditions.
Lipid and energy metabolism
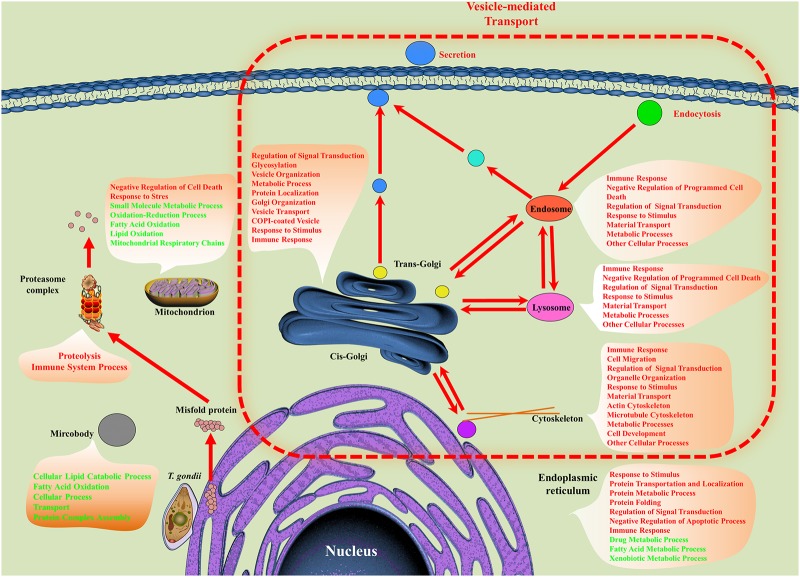
As shown in Fig 4, some biological processes or functions of hepatocellular organelles were altered. For example, negative regulation of cell death was upregulated in lysosomes, endosomes, mitochondria and ER, which is consistent with the anti-apoptotic activity of T. gondii [49]. In the mitochondria of hepatocytes, energy production was decreased by downregulation of mitochondrial respiratory chain, lipid oxidation, and fatty acid oxidation. Lipid catabolic processes and fatty acid oxidation were downregulated in microbodies too. Mitochondria, lipids or fatty acids are important sources of energy, which is needed for many biological processes. These downregulations imply that hepatocellular energy metabolism was affected by T. gondii infection. Severe alterations of energy production in mitochondria could result in several liver diseases such as liver necrosis, steatosis or steatohepatitis [50]. These diseases are predicted to be linked to the DEPs obtained in the present study as indicated by the Comparative Toxicogenomics Database analysis.
Fig 4. Differentially expressed GO term of cellular organelle.
The processes, such as secretion, endocytosis and vesicle-mediated transport were enriched by all DEPs. GO term of cellular organelles were enriched using differentially expressed cellular organelle components. All enrichment analysis was performed on BiNGO plugin. Red color represents upregulated GO term. Green color represents downregulated GO term.
Drug detoxification
The liver is the major detoxifying organ in the mammalian body. Its ability to clear most drugs and other xenobiotics determine the efficacy and toxicity associated with any given drug. Previous reports showed that T. gondii infection can alter drug pharmacokinetics too [10,11]. In general, xenobiotic or drugs metabolism in vivo occurs in three phases [51]. Phase-I is mainly mediated by the members of cytochrome P450 family which transform lipophilic materials into soluble hydrophilic substrates. The products of phase-I are toxic or electrophilic intermediates and if these products not transformed into nontoxic substances by the metabolic process of phase-II, cancer or tissue damage could be induced [52]. The phase-II reactions are mediated by several transferases or oxidoreductase, such as UDP glucuronyltransferases, glutathione-S-transferases, sulfotransferases, alcohol dehydrogenase, aldo-keto reductase, flavin containing monooxygenase and other enzymes involved in xenobiotic or drug metabolic processes. In phase-III, all products that are detoxified by phase-II reactions are excreted into bile via export pumps and can then be eliminated from the body. In the present study, one glutathione S-transferase (Gstm1), two sulfotransferases (Sult2a1 and Sult2a5), eleven cytochrome P450 (Cyp2c54, Cyp3a16, Cyp3a11, Cyp8b1, Cyp2c50, Cyp2b9, Cyp4a10, Cyp3a25, Cyp1a2, Cyp2a22, and Cyp2c29) and one bile salt export pump (Abcb11) were downregulated. Only one cytochrome P450 (Cyp3a13) was upregulated in infected liver.
Downregulated metabolisms of macrolide antibiotics and cyclosporin A, which are listed in S5 Table, have been observed during T. gondii infection [10,11]. Previous reports showed that the altered pharmacokinetics of macrolide antibiotics or cyclosporin could result in severe adverse drug reactions [53,54]. Therefore, knowledge of the pharmacokinetics changes that occur in toxoplasmosis is very important for drug prescription purposes particularly when multiple drugs or doses are taken in conjunction. According to the analysis of the Comparative Toxicogenomics Database, the metabolism of over 400 substances were altered in infected liver, including that of antibiotics, pesticides, tranquilizer, glucocorticoids, and other xenobioticsor/drugs. The top 10 substances that their metabolism could be affected by T. gondii infection were 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene, dietary fats, phenobarbital, perfluorooctanoic acid, pirinixic acid, benzo(a)pyrene, diethylnitrosamine, tetrachlorodibenzodioxin, clofibrate, and acetaminophen. Acetaminophen (also known as N-acetyl-p-aminophenol or paracetamol) is a commonly used analgesic and antipyretic drug that can cause extensive hepatic damage after an excessive dose and is the leading cause of drug-induced liver injury in the USA [55]. Acetaminophen-mediated hepatotoxicity has been thought to be attributed to an initial oxidative stress followed by energetic stress, mitochondrial dysfunction and upregulation of glycolysis [56]. Similar metabolic alterations have been observed during T. gondii infection [57]. Therefore, failure to take these changes into consideration when acetaminophen is prescribed to toxoplasmosis patients can result in increased risk of developing severe acetaminophen-induced acute liver damage. T. gondii infection will not only potentiate the oxidative and bioenergetic stress caused by acetaminophen, but will also change the drug pharmacokinetic via downregulation of the drug metabolic processes It is possible that the xenobiotic or drugs metabolics changes are not specific to T. gondii infection. However, our data clearly indicate that proteomic alterations are not global, but rather target specific cellular functions, such as the metabolism of the xenobiotic/drugs metabolism in mouse liver following acute T. gondii infection. Also, the mouse model and approach used in our study can be useful in elucidating the pharmacokinetic changes of any given compound before being prescribed to toxoplasmosis patients.
In summary, we profiled, for the first time, the mouse liver proteome following acute T. gondii infection at a global level. Our findings demonstrate that T. gondii infection has wide spread effects on the expression of host proteins involved in host immune defense mechanisms, lipid metabolism, intracellular transport, and drug pharmacokinetics. The results of this research lead to an important question: could the interaction of T. gondii with liver tissue impair mitochondrial functions and liver metabolism to such a level that T. gondii-induced hepatic dysfunction might be explained? There is evidence that T. gondii infection is associated with extensive alterations in the liver of affected hosts [3–6,8–11]. The biomarker molecules and pathways identified in our study also provide insights into some of the underlying causes of T. gondii-associated liver pathology. Inefficient mitochondrial metabolism can affect hepatocellular energy metabolism and could result in several liver diseases such as liver necrosis, steatosis or steatohepatitis [50], highlighting the importance of further efforts to understand the interaction of T. gondii with mitochondria and its influence on hepatic metabolism. While hundreds of differentially expressed proteins are observed in our study, the validation and clinical value of these proteins warrants further investigation.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
BGI-Shenzhen is thanked for technical assistance. The authors thank Dr. Alasdair Nisbet, Vaccines and Diagnostics, Moredun Research Institute, Scotland, UK for help in improving the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Project support was provided by the National Natural Science Foundation of China (Grant No. 31230073) and the Science Fund for Creative Research Groups of Gansu Province (Grant No. 1210RJIA006). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sibley L (2004) Intracellular parasite invasion strategies. Science 304: 248–253. [DOI] [PubMed] [Google Scholar]
- 2.Iaccheri B, Fiore T, Papadaki T, Androudi S, Janjua S, Bhaila I, et al. (2008) Adverse drug reactions to treatments for ocular toxoplasmosis: a retrospective chart review. Clin Ther 30: 2069–2074. 10.1016/j.clinthera.2008.10.021 [DOI] [PubMed] [Google Scholar]
- 3.Weitberg AB, Alper JC, Diamond I, Fligiel Z (1979) Acute granulomatous hepatitis in the course of acquired toxoplasmosis. N Engl J Med 300: 1093–1096. [DOI] [PubMed] [Google Scholar]
- 4.Tiwari I, Rolland C, Popple A (1982) Cholestatic jaudice due to Toxoplasma hepatitis. Postgrad Med J 58: 299–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ustun S, Aksoy U, Dagci H, Ersoz G (2004) Frequency of toxoplasmosis in patients with cirrhosis. World J Gastroenterol 10: 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shapira Y, Agmon-Levin N, Renaudineau Y, Porat-Katz BS, Barzilai O, Ram M, et al. (2012) Serum markers of infections in patients with primary biliary cirrhosis: evidence of infection burden. Exp Mol Pathol 93: 386–390. 10.1016/j.yexmp.2012.09.012 [DOI] [PubMed] [Google Scholar]
- 7.Mordue DG, Monroy F, La Regina M, Dinarello CA, Sibley LD (2001) Acute toxoplasmosis leads to lethal overproduction of Th1 cytokines. J Immunol 167: 4574–4584. [DOI] [PubMed] [Google Scholar]
- 8.Wendum D, Carbonell N, Svrcek M, Chazouilléres O, Flejou J (2002) Fatal disseminated toxoplasmosis in a toxoplasma seropositive liver transplant recipient. J Clin Pathol 55: 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atmaca HT, Gazyagcı AN, Canpolat S, Kul O (2013) Hepatic stellate cells increase in Toxoplasma gondii infection in mice. Parasites Vectors 6: 135 10.1186/1756-3305-6-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg-Candolfi M, Candolfi E (1996) Depression of the N-demethylation of erythromycin, azithromycin, clarithromycin and clindamycin in murine Toxoplasma infection. Int J Parasitol 26: 1321–1323. [DOI] [PubMed] [Google Scholar]
- 11.Berg-Candolfi M, Candolfi E, Benet L (1996) Suppression of intestinal and hepatic cytochrome P4503A in murine Toxoplasma infection. Effects of N-acetylcysteine and N G-monomethyl-L-arginine on the hepatic suppression. Xenobiotica 26: 381–394. [DOI] [PubMed] [Google Scholar]
- 12.Giannini EG, Testa R, Savarino V (2005) Liver enzyme alteration: a guide for clinicians. CMAJ 172: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson M, Jones A, Carmen J, Sinai A, Burchmore R, Wastling J (2008) Modulation of the host cell proteome by the intracellular apicomplexan parasite Toxoplasma gondii. Infect Immun 76: 828–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou DH, Yuan ZG, Zhao FR, Li HL, Zhou Y, Lin RQ, et al. (2011) Modulation of mouse macrophage proteome induced by Toxoplasma gondii tachyzoites in vivo. Parasitol Res 109: 1637–1646. 10.1007/s00436-011-2435-z [DOI] [PubMed] [Google Scholar]
- 15.Zhou DH, Zhao FR, Huang SY, Xu MJ, Song HQ, Su C, et al. (2013) Changes in the proteomic profiles of mouse brain after infection with cyst-forming Toxoplasma gondii. Parasit Vectors 6: 96 10.1186/1756-3305-6-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melo MB, Nguyen QP, Cordeiro C, Hassan MA, Yang N, McKell R, et al. (2013) Transcriptional analysis of murine macrophages infected with different Toxoplasma strains identifies novel regulation of host signaling pathways. PLoS Pathog 9: e1003779 10.1371/journal.ppat.1003779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He JJ, Ma J, Song HQ, Zhou DH, Wang JL, Huang SY, et al. (2015) Transcriptomic analysis of global changes in cytokine expression in mouse spleens following acute Toxoplasma gondii infection. Parasitol Res: 1–10. [DOI] [PubMed] [Google Scholar]
- 18.Tymoshenko S, Oppenheim RD, Agren R, Nielsen J, Soldati-Favre D, Hatzimanikatis V (2015) Metabolic Needs and Capabilities of Toxoplasma gondii through Combined Computational and Experimental Analysis. PLoS Comput Biol 11: e1004261 10.1371/journal.pcbi.1004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou CX, Zhou DH, Elsheikha HM, Liu GX, Suo X, Zhu XQ (2015) Global Metabolomic Profiling of Mice Brains following Experimental Infection with the Cyst-Forming Toxoplasma gondii. PLoS One 10: e0139635 10.1371/journal.pone.0139635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang HH, Huang SY, Zhou DH, Zhang XX, Su C, Deng SZ, et al. (2013) Genetic characterization of Toxoplasma gondii from pigs from different localities in China by PCR-RFLP. Parasites Vectors 6: 227 10.1186/1756-3305-6-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 22.Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis AP, Grondin CJ, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, et al. (2015) The Comparative Toxicogenomics Database's 10th year anniversary: update 2015. Nucleic Acids Res 43: D914–D920. 10.1093/nar/gku935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zambelli F, Pesole G, Pavesi G (2009) Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res 37: W247–W252. 10.1093/nar/gkp464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, et al. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32: 223–226. 10.1038/nbt.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blader IJ, Saeij JP (2009) Communication between Toxoplasma gondii and its host: impact on parasite growth, development, immune evasion, and virulence. APMIS 117: 458–476. 10.1111/j.1600-0463.2009.02453.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yap GS, Sher A (1999) Effector cells of both nonhemopoietic and hemopoietic origin are required for interferon (IFN)-γ–and tumor necrosis factor (TNF)-α–dependent host resistance to the intracellular pathogen, Toxoplasma gondii. J Exp Med 189: 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yarovinsky F (2014) Innate immunity to Toxoplasma gondii infection. Nat Rev Immunol 14: 109–121. 10.1038/nri3598 [DOI] [PubMed] [Google Scholar]
- 31.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem 389: 1017–1031. [DOI] [PubMed] [Google Scholar]
- 32.Zeng J, Parvanova IA, Howard JC (2009) A dedicated promoter drives constitutive expression of the cell-autonomous immune resistance GTPase, Irga6 (IIGP1) in mouse liver. PLoS One 4: e6787 10.1371/journal.pone.0006787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melzer T, Duffy A, Weiss L, Halonen S (2008) The gamma interferon (IFN-γ)-inducible GTP-binding protein IGTP is necessary for Toxoplasma vacuolar disruption and induces parasite egression in IFN-γ-stimulated astrocytes. Infect Immun 76: 4883–4894. 10.1128/IAI.01288-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degrandi D, Konermann C, Beuter-Gunia C, Kresse A, Würthner J, Kurig S, et al. (2007) Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J Immunol 179: 7729–7740. [DOI] [PubMed] [Google Scholar]
- 35.Fentress SJ, Behnke MS, Dunay IR, Mashayekhi M, Rommereim LM, Fox BA, et al. (2010) Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe 8: 484–495. 10.1016/j.chom.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahmoud ME, Ui F, Salman D, Nishimura M, Nishikawa Y (2015) Mechanisms of interferon-beta-induced inhibition of Toxoplasma gondii growth in murine macrophages and embryonic fibroblasts: role of immunity-related GTPase M1. Cell Microbiol 17: 1069–1083. 10.1111/cmi.12423 [DOI] [PubMed] [Google Scholar]
- 37.Sturge CR, Benson A, Raetz M, Wilhelm CL, Mirpuri J, Vitetta ES, et al. (2013) TLR-independent neutrophil-derived IFN-gamma is important for host resistance to intracellular pathogens. Proc Natl Acad Sci U S A 110: 10711–10716. 10.1073/pnas.1307868110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arend WP, Malyak M, Guthridge CJ, Gabay C (1998) Interleukin-1 receptor antagonist: role in biology. Annu Rev Immunol 16: 27–55. [DOI] [PubMed] [Google Scholar]
- 39.Lüder CG, Walter W, Beuerle B, Maeurer MJ, Gross U (2001) Toxoplasma gondii down-regulates MHC class II gene expression and antigen presentation by murine macrophages via interference with nuclear translocation of STAT1α. Eur J Immunol 31: 1475–1484. [DOI] [PubMed] [Google Scholar]
- 40.Shapira S, Speirs K, Gerstein A, Caamano J, Hunter C (2002) Suppression of NF-κB activation by infection with Toxoplasma gondii. J Infect Dis 185: S66–S72. [DOI] [PubMed] [Google Scholar]
- 41.Kim S-K, Fouts AE, Boothroyd JC (2007) Toxoplasma gondii dysregulates IFN-γ-inducible gene expression in human fibroblasts: insights from a genome-wide transcriptional profiling. J Immunol 178: 5154–5165. [DOI] [PubMed] [Google Scholar]
- 42.Lang C, Hildebrandt A, Brand F, Opitz L, Dihazi H, Lüder CG (2012) Impaired Chromatin Remodelling at STAT1-Regulated Promoters Leads to Global Unresponsiveness of Toxoplasma gondii-Infected Macrophages to IFN-γ. PLoS Pathog 8: e1002483 10.1371/journal.ppat.1002483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butcher BA, Kim L, Panopoulos AD, Watowich SS, Murray PJ, Denkers EY (2005) Cutting edge: IL-10-independent STAT3 activation by Toxoplasma gondii mediates suppression of IL-12 and TNF-α in host macrophages. J Immunol 174: 3148–3152. [DOI] [PubMed] [Google Scholar]
- 44.Stutz A, Kessler H, Kaschel M-E, Meissner M, Dalpke AH (2012) Cell invasion and strain dependent induction of suppressor of cytokine signaling-1 by Toxoplasma gondii. Immunobiology 217: 28–36. 10.1016/j.imbio.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitmarsh RJ, Gray CM, Gregg B, Christian DA, May MJ, Murray PJ, et al. (2011) A critical role for SOCS3 in innate resistance to Toxoplasma gondii. Cell Host Microbe 10: 224–236. 10.1016/j.chom.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pryer NK, Wuestehube LJ, Schekman R (1992) Vesicle-mediated protein sorting. Annu Rev Biochem 61: 471–516. [DOI] [PubMed] [Google Scholar]
- 47.Blum JS, Wearsch PA, Cresswell P (2013) Pathways of antigen processing. Annu Rev Immunol 31: 443 10.1146/annurev-immunol-032712-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka S, Nishimura M, Ihara F, Yamagishi J, Suzuki Y, Nishikawa Y (2013) Transcriptome analysis of mouse brain infected with Toxoplasma gondii. Infect Immun 81: 3609–3619. 10.1128/IAI.00439-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lüder C, Gross U (2005) Apoptosis and its modulation during infection with Toxoplasma gondii: molecular mechanisms and role in pathogenesis. Curr Top Microbiol Immunol 289: 219–237. [DOI] [PubMed] [Google Scholar]
- 50.Pessayre D, Fromenty B, Berson A, Robin M-A, Lettéron P, Moreau R, et al. (2012) Central role of mitochondria in drug-induced liver injury. Drug Metab Rev 44: 34–87. 10.3109/03602532.2011.604086 [DOI] [PubMed] [Google Scholar]
- 51.Xu C, Li CY-T, Kong A-NT (2005) Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res 28: 249–268. [DOI] [PubMed] [Google Scholar]
- 52.Talalay P (2000) Chemoprotection against cancer by induction of phase 2 enzymes. Biofactors 12: 5–12. [DOI] [PubMed] [Google Scholar]
- 53.Periti P, Mazzei T, Mini E, Novelli A (1993) Adverse effects of macrolide antibacterials. Drug Saf 9: 346–364. [DOI] [PubMed] [Google Scholar]
- 54.Scott J, Higenbottam T (1988) Adverse reactions and interactions of cyclosporin. Med Toxicol Adverse Drug Exp 3: 107–127. [DOI] [PubMed] [Google Scholar]
- 55.Larson AM, Polson J, Fontana RJ, Davern TJ, Lalani E, Hynan LS, et al. (2005) Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology 42: 1364–1372. [DOI] [PubMed] [Google Scholar]
- 56.Kyriakides M, Maitre L, Stamper BD, Mohar I, Kavanagh TJ, Foster J, et al. (2016) Comparative metabonomic analysis of hepatotoxicity induced by acetaminophen and its less toxic meta-isomer. Arch Toxicol: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menendez MT, Teygong C, Wade K, Florimond C, Blader IJ (2015) siRNA Screening Identifies the Host Hexokinase 2 (HK2) Gene as an Important Hypoxia-Inducible Transcription Factor 1 (HIF-1) Target Gene in Toxoplasma gondii-Infected Cells. mBio 6: e00462–00415. 10.1128/mBio.00462-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.