Fig. 4.
Comparison between targeted exome and whole exome approaches.
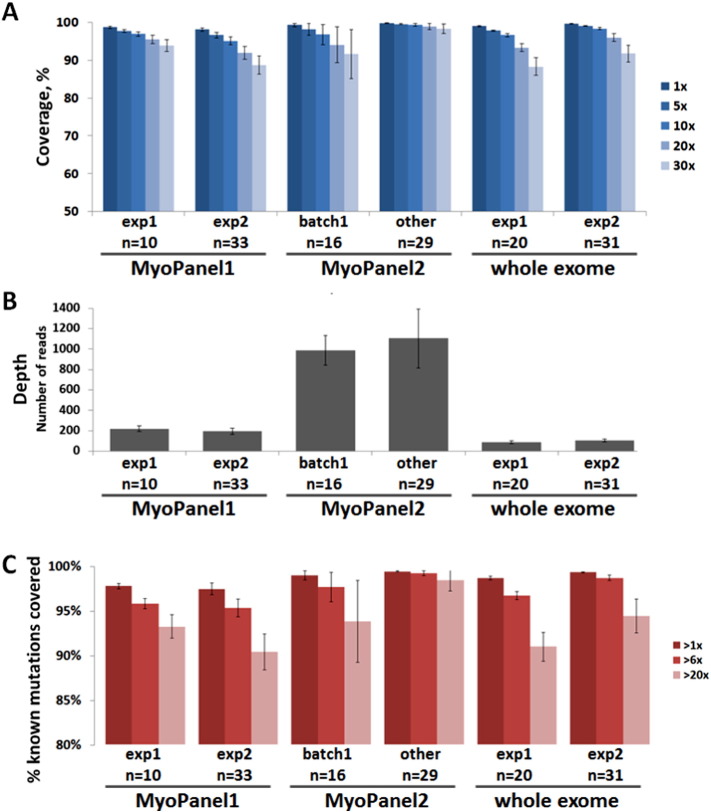
A. Percentage of target region coverage for all sequencing experiments. A set of 5817 unique exons, corresponding to 295 genes found in both MyoPanel1 and MyoPanel2, was used for coverage and depth analysis. Average sequence coverage at 20 × was 95.5% for MyoPanel1-exp1, 92% for MyoPanel1-exp2, 94.1% for lower-quality (batch1) MyoPanel2 samples, 98.8% for other MyoPanel2 samples, 93.4% and 96% for two different whole exome sequencing experiments. B. Depth of coverage for all sequencing experiments. Average depth of coverage was 221 for MyoPanel1-exp1, 195 for MyoPanel1-exp2, 989 for lower-quality (batch1) MyoPanel2 samples, 1106 for other MyoPanel2 samples, 88 and 104 for two different whole exome sequencing experiments. C. Percentage of known mutation positions detected by sequencing test. A set of 11,467 published mutations was used for calculations. Average number of mutation positions covered more than 6 × was 96.2% for MyoPanel1-exp1, 95.7% for MyoPanel1-exp2, 98.1% for lower-quality (batch1) MyoPanel2 samples, 99.7% for other MyoPanel2 samples, 97.1% and 99.2% for two different whole exome sequencing experiments. Averages ± SD are shown.