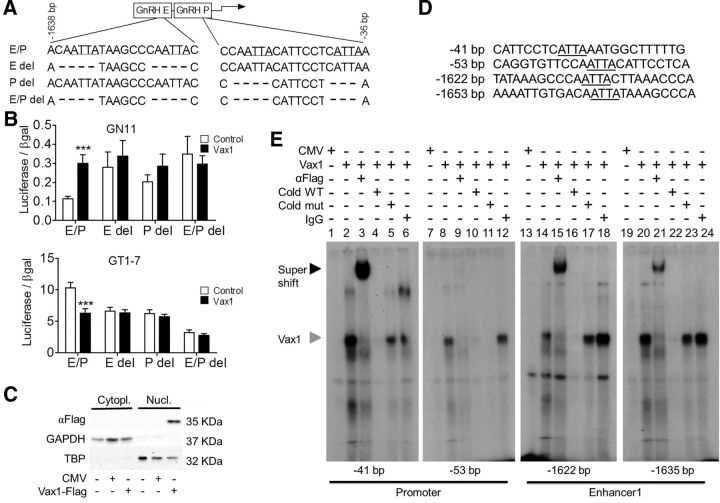
Figure 6.
VAX1 regulates GnRH transcription through direct binding to ATTA sites in the GnRH-E1 and proximal promoter (P). A, Deletion of potential ATTA binding sites of VAX1 within the GnRH enhancer1/promoter (GnRH-E/P or E/P). B, Cotransfection of the indicated luciferase reporters with (black) or without (white) 10 ng of Vax1 in GN11 and GT1-7 cells. Data are expressed as fold-induction compared with GnRH E/P. Statistical analysis by two-way ANOVA followed by Bonferroni. ***p < 0.001 compared with control. C, COS-1 cells were transiently transfected with a Vax1-flag expression vector or its empty vector, CMV, and harvested 24 h post-transfection. Western blot shows detection of VAX1-flag, GAPDH (glyceraldehyde-3phosphate dehydrogenase), and TBP in the nucleus (Nucl) versus the cytoplasm (Cytopl). D, Probe sequences used for EMSA covering the ATTA sites found to be involved in VAX1 regulation of Gnrh1 transcription (sites indicated in A). The underlined ATTA elements were converted to GCCG in the mutant probes (Cold mut). E, EMSA performed using nuclear extracts from COS-1 cells transfected with Vax1-flag plasmid or control expression vector. Black arrowhead indicates super shift, gray arrowhead indicates origin of supershifted band. Wild-type (Cold WT) and mutant (Cold mut) probes were used at 100-fold higher concentration than labeled WT probe.