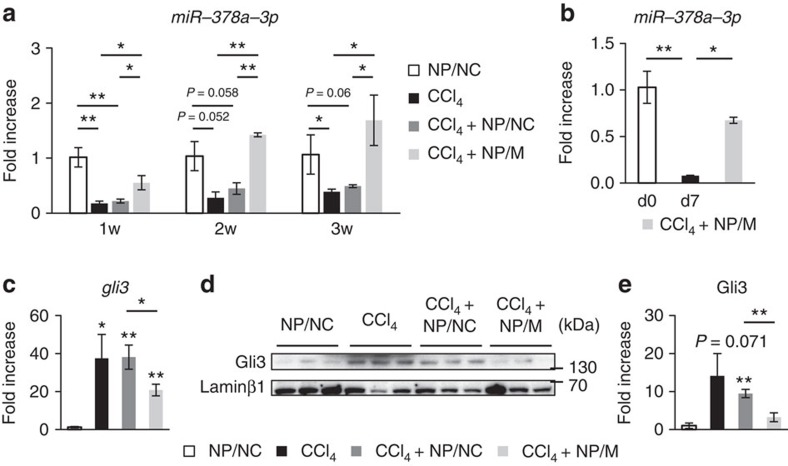
Figure 7. NPs having miR-378a-3p increase the level of miR-378a-3p but decrease Gli3 expression in CCl4-treated mice.
(a) qRT–PCR for miR-378a-3p in livers from NP/NC, CCl4, CCl4-treated with NP/NC (CCl4+NP/NC) or NP/M (CCl4+NP/M) mice (n=4 per group). Mean±s.e.m. results are graphed (Kruskal–Wallis test and unpaired two-sample Student's t-test, *P<0.05 and **P<0.005 versus NP/NC). (b) qRT–PCR of miR-378a-3p in primary qHSCs isolated from normal mice at quiescent stage (d0; immediately after isolation), primary aHSCs (d7; cultured for 7 days) and primary HSCs isolated from CCl4+NP/M mice at 3 weeks after NPs treatment. Results of relative expression values are shown as mean±s.e.m. of triplicate experiments (one-way analysis of variance (ANOVA) with Tukey corrections, *P<0.05 and **P<0.005 versus d0). (c) qRT–PCR analysis for gli3 in livers from all mice at 3 weeks after NPs treatment (n=4 per group). Mean±s.e.m. results are graphed (Kruskal–Wallis test and unpaired two-sample Student's t-test, *P<0.05 and **P<0.005 versus NP/NC). (d) Western blot analysis for Gli3 (145 kDa) and Laminβ1 (68 kDa, internal control for nuclear fraction) in livers of three representative mice from each group. Data shown represent one of three experiments with similar results. (e) Cumulative densitometric analyses of Gli3 western blotting results are displayed as the mean±s.e.m. (Kruskal–Wallis test and unpaired two-sample Student's t-test, **P<0.05).