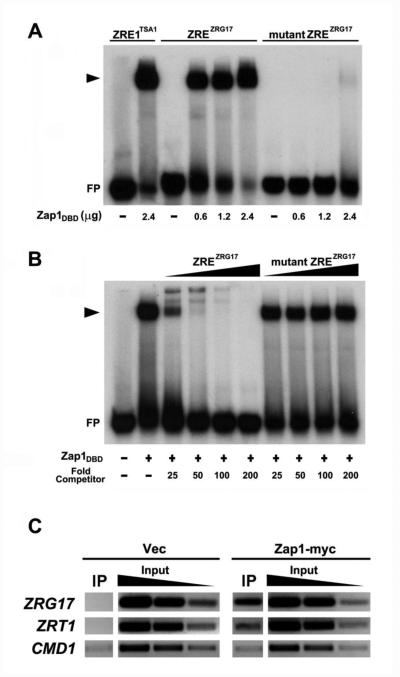
Figure 4. Zap1 binds specifically to the ZRG17 ZRE in vitro and in vivo.
A) Electrophoretic mobility shift assay (EMSA) showing sequence specificity of Zap1 DNA-binding domain (Zap1DBD) binding to the ZRG17 ZRE. Radiolabeled oligonucleotides with the wild-type TSA1 ZRE1 (ZRE1TSA1), wild-type ZRG17 ZRE (ZREZRG17), or the mutated ZRG17 m1ZRE (mutant ZREZRG17) were incubated with purified Zap1DBD for 1 hour prior to native gel electrophoresis. The experiments were performed with the indicated amounts of Zap1DBD. FP denotes the free probe, and the arrowhead indicates the Zap1DBD-ZRE complex. B) EMSA competition assay showing that the ZRG17 ZRE competes with TSA1 ZRE1 for Zap1DBD binding. Radiolabeled wild-type TSA1 ZRE1 (ZRE1TSA1) oligonucleotides were incubated with 0 (−) or 2.4 μg (+) Zap1pDBD and a 25-, 50-, 100-, or 200-fold excess of the nonradiolabeled wild-type ZRG17 ZRE (ZREZRG17) or the mutated ZRG17 m1ZRE (mutant ZREZRG17) oligonucleotides for 1 h at room temperature prior to native gel electrophoresis. C) Chromatin immunoprecipitation verifies Zap1 binding to the ZRG17 promoter in vivo. Wild-type cells transformed with either the vector (pYef2L) or a plasmid expressing a myc-tagged Zap1 protein (Zap1-myc) were grown under low zinc conditions (LZM + 3 μM ZnCl2). Chromatin immunoprecipitation was performed as described in the Experimental section. Enrichment of the ZRG17 ZRE was observed in the immunoprecipitates (IP) from Zap1-myc expressing cells relative to the vector-only cells. The ZRT1 ZRE and CMD1 promoter regions were used as positive and negative controls, respectively. PCR products generated from 10-fold serially diluted input samples were used to confirm the semi-quantitativeness of the analysis. The data shown are representative of two independent experiments.