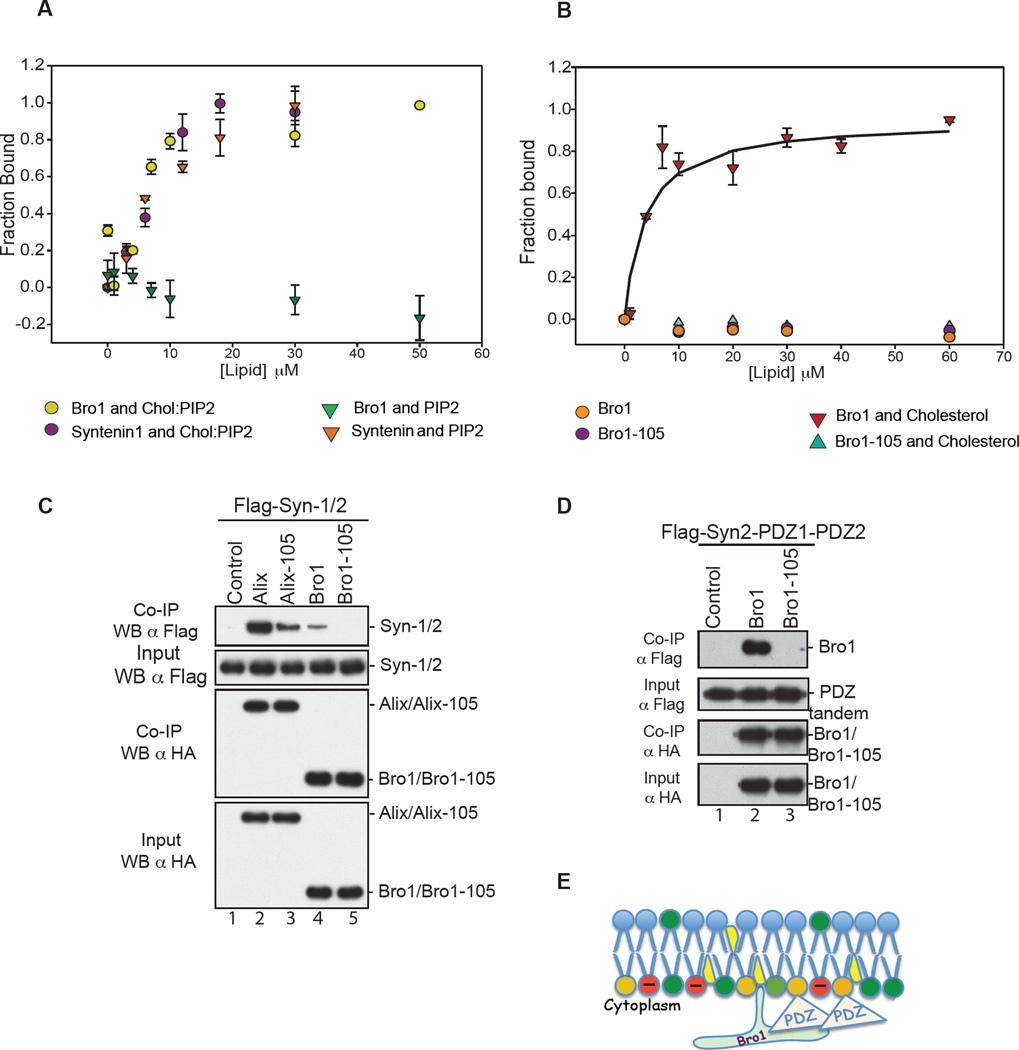
Figure 2.
(A) ALIX Bro1 binds the membrane in PI(4,5)P2-independent manner. Comparison of membrane binding of 130 nM CPM-Bro1 ( ) and 130 nM Syn-1 (
) and 130 nM Syn-1 ( ) to LUVs containing both PI(4,5)P2 and cholesterol, (PC:PE:PS:Chol: PI(4,5)P2 at 26:33:27:9:5) (n=6), to membrane binding of CPM-Bro1 (
) to LUVs containing both PI(4,5)P2 and cholesterol, (PC:PE:PS:Chol: PI(4,5)P2 at 26:33:27:9:5) (n=6), to membrane binding of CPM-Bro1 ( ) and CPM-Syn-1 (
) and CPM-Syn-1 ( ) to membranes without cholesterol, (PC:PS:PE: PI(4,5)P2 at 28:34:28:10), (n=6). (B) Cholesterol is critical for ALIX Bro1 binding to the membrane. The fraction of protein bound to membranes was determined by the increase in fluorescence of a 130 nM CPM-Bro1 or the CPM-Bro1 105 mutant as LUVs with or without cholesterol were incrementally added. Neither CPM-Bro1 (
) to membranes without cholesterol, (PC:PS:PE: PI(4,5)P2 at 28:34:28:10), (n=6). (B) Cholesterol is critical for ALIX Bro1 binding to the membrane. The fraction of protein bound to membranes was determined by the increase in fluorescence of a 130 nM CPM-Bro1 or the CPM-Bro1 105 mutant as LUVs with or without cholesterol were incrementally added. Neither CPM-Bro1 ( ) nor CPM-Bro1-105 (
) nor CPM-Bro1-105 ( ) bound to PC:PE:PS (1:1:1) lipids (n=6), while CPM-Bro1 (
) bound to PC:PE:PS (1:1:1) lipids (n=6), while CPM-Bro1 ( ) but not CPM-Bro1-105 (
) but not CPM-Bro1-105 ( ) showed significant binding to PC:PE:PS:Cholesterol (28:34:28:10) lipids (n=9). (C) Flag- Syntenin expressed in 293T cells alone (lane 1), or with either HA-Alix (lane 2), HA-Alix-105 (lane 3), HA-Bro1 (lane 4) or HA-Bro1-105 (lane 5). (D) In parallel, cells were also transfected with Flag-Syn-2 PDZ tandem (1/2) alone (lane 1) or with HA-Bro1 (lane 2) or HA-Bro1 Phe105 (lane 3). Cell lysates were incubated with anti-HA antibody-conjugated beads. Both input and IP complexes were analyzed by SDS-PAGE and WB as indicated. (E) Model of ALIX Bro1-syn-2 PDZ tandem co-anchorage at lipid-rich domains of the membrane. PI(4,5)P2 (orange), cholesterol (yellow), anionic phospholipids PS (red), Bro1 and Phe105 loop (green), and PDZs tandem (pink). See also S3.
) showed significant binding to PC:PE:PS:Cholesterol (28:34:28:10) lipids (n=9). (C) Flag- Syntenin expressed in 293T cells alone (lane 1), or with either HA-Alix (lane 2), HA-Alix-105 (lane 3), HA-Bro1 (lane 4) or HA-Bro1-105 (lane 5). (D) In parallel, cells were also transfected with Flag-Syn-2 PDZ tandem (1/2) alone (lane 1) or with HA-Bro1 (lane 2) or HA-Bro1 Phe105 (lane 3). Cell lysates were incubated with anti-HA antibody-conjugated beads. Both input and IP complexes were analyzed by SDS-PAGE and WB as indicated. (E) Model of ALIX Bro1-syn-2 PDZ tandem co-anchorage at lipid-rich domains of the membrane. PI(4,5)P2 (orange), cholesterol (yellow), anionic phospholipids PS (red), Bro1 and Phe105 loop (green), and PDZs tandem (pink). See also S3.