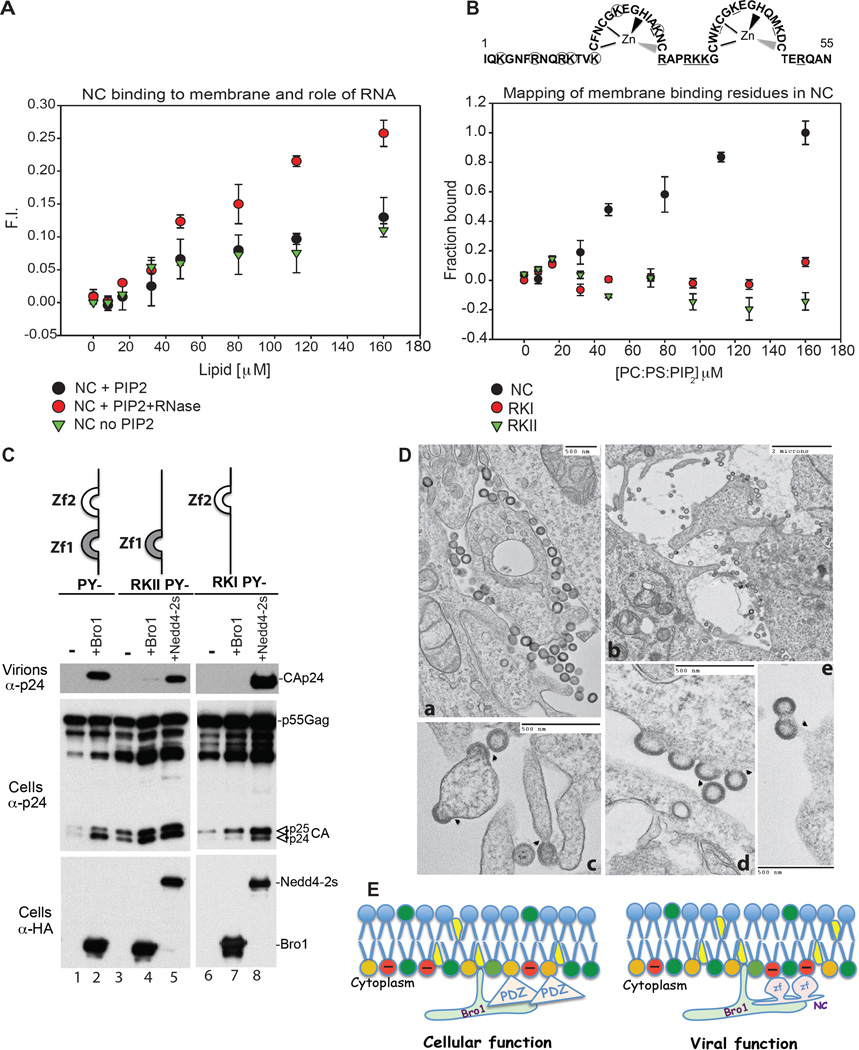
Figure 3.
(A) NC binding to membrane is enhanced by RNAse treatment. The fraction of NC bound to lipid was determined by the increase in fluorescence of the CPM-labeled protein as LUVs were incrementally added. Binding of 130 nM CPM-NC treated with RNAse to LUVs containing PI(4,5)P2 (PC:PE:PS: PI(4,5)P2 at 31:32:32:5) ( ) showed a different pattern when compared to NC binding to LUVs with PI(4,5)P2 PC:PE:PS: PI(4,5)P2 at 31:32:32:5) without RNAse (
) showed a different pattern when compared to NC binding to LUVs with PI(4,5)P2 PC:PE:PS: PI(4,5)P2 at 31:32:32:5) without RNAse ( ) or without PI(4,5)P2 (PC:PE:PS at 1:1:1) (
) or without PI(4,5)P2 (PC:PE:PS at 1:1:1) ( ) (n=9). (B) Disruption of NC zinc finger basic residues ablates membrane binding. Substitution to A in RKI (circles) or RKII (underlined) are shown above the panel. While 130 nM CPM-WT NC was observed to bind to LUVs containing PI(4,5)P2 (PC:PE:PS: PI(4,5)P2 at 31:32:32:5) (
) (n=9). (B) Disruption of NC zinc finger basic residues ablates membrane binding. Substitution to A in RKI (circles) or RKII (underlined) are shown above the panel. While 130 nM CPM-WT NC was observed to bind to LUVs containing PI(4,5)P2 (PC:PE:PS: PI(4,5)P2 at 31:32:32:5) ( ), neither 130 nM CPM-NC RKI (
), neither 130 nM CPM-NC RKI ( ) nor 130 nM CPM-NC RKII (
) nor 130 nM CPM-NC RKII ( ) bound to the same lipid (n=9) in identical conditions. (C) Disruption of NC membrane binding ability prevents utilization of ALIX Bro1 in HIV-1 budding. 293T cells were transfected with HIV-1 PY- (lanes 1–2), RKIIPY- (3–5) or RKIPY- (lanes 6–8) either alone, with HA-Bro1 (lanes 2, 4 and 7) or HA-Nedd4.2s (lanes 5 and 8). Cells and virus protein content was analyzed by SDS-PAGE and WB. (D) TEM images of RKI (a, d, e) and RKII (b, c) reveal arrested particles at the PM. (E) Model of ALIX Bro1-HIV-1 NC co-anchorage at lipid-rich domains for viral and cellular function. PI(4,5)P2 (orange), zinc fingers (zf) (pink).
) bound to the same lipid (n=9) in identical conditions. (C) Disruption of NC membrane binding ability prevents utilization of ALIX Bro1 in HIV-1 budding. 293T cells were transfected with HIV-1 PY- (lanes 1–2), RKIIPY- (3–5) or RKIPY- (lanes 6–8) either alone, with HA-Bro1 (lanes 2, 4 and 7) or HA-Nedd4.2s (lanes 5 and 8). Cells and virus protein content was analyzed by SDS-PAGE and WB. (D) TEM images of RKI (a, d, e) and RKII (b, c) reveal arrested particles at the PM. (E) Model of ALIX Bro1-HIV-1 NC co-anchorage at lipid-rich domains for viral and cellular function. PI(4,5)P2 (orange), zinc fingers (zf) (pink).