Fig. 3.
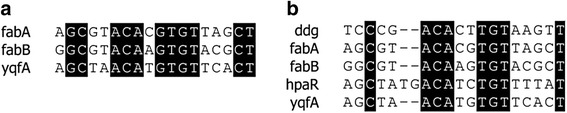
Alignment of the putative FabR binding site in the ChIP-qPCR verified in vivo FabR targets. a Alignment of the already known E. coli FabR targets [51] in S. Typhimurium SL1344; b Alignment of the ChIP-chip identified and ChIP-qPCR verified FabR targets in S. Typhimurium SL1344. All alignments were performed using MotifSampler [47]. Sequences upstream of the coding sequences of the indicated genes were taken from the complete genome sequence of S. Typhimurium SL1344. These input sequences comprised the full intergenic region, i.e. the region between the coding sequence of the FabR target gene and the upstream coding sequence, with hpaR and ddg indicating the hpaR/hpaG and ddg/yfdZ intergenic sequences, respectively. White letters with black background denote identical bases and black letters on a white background denote differing bases
