Extended Data Figure 4. Building an atomic model of InsP3R1.
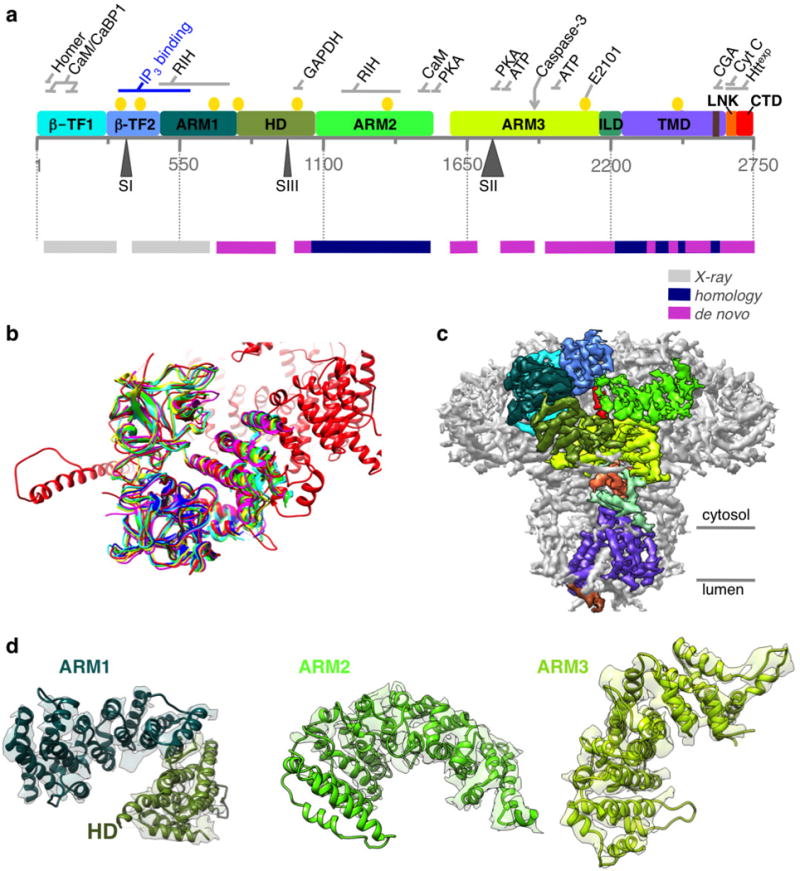
a, Linear representation of a primary structure of InsP3R1 protein (GI accession 17380349). Ten domains identified in the cryo-EM density map are colour-coded. Three sites of alternative splicing (residues 318–332/SI, 918–926/SIII and 1692–1731/SII) are indicated below the sequence bar. Putative binding sites for several channel-specific ligands are indicated above the domains (ATP, ATP-binding CaM/CaBP, calmodulin/Ca2+ binding protein; CGA, chromogranin A; cyt c, cytochrome c; Httexp, huntingtin; PKA, protein kinase A; RIH, RyR/InsP3R homology; yellow circles denote Ca2+-binding20). The panel below shows a linear diagram of the protein sequence colour-coded based on the approach used for modelling different regions in the primary structure. The spaces between the bars correspond to unmodelled sequence (see also Extended Data Fig. 5). b, The structure of the N-terminal domain (NTD) based on cryo-EM density map (red) is shown overlapped with the X-ray crystal structures of NTD (r.m.s.d. values 5 1.3–1.4 Å): 1XZZ (blue); 3T8Sa (cyan); 3T8Sb (green); 3UJ4 (yellow); and 3UJ0 (magenta). c, Cryo-EM density map is viewed along the membrane plane. Densities corresponding to the individual domains of one subunit of InsP3R1 are colour-coded as in a. d, Models for solenoid-like α-helical domains are shown superimposed on their corresponding cryo-EM densities. ARM1–ARM3, armadillo repeat domains1–3; HD, α-helical domain. The densities of ARM2 domain are less resolved than those of ARM1 and ARM3 but are sufficient to trace the backbone.
