Fig. 1.
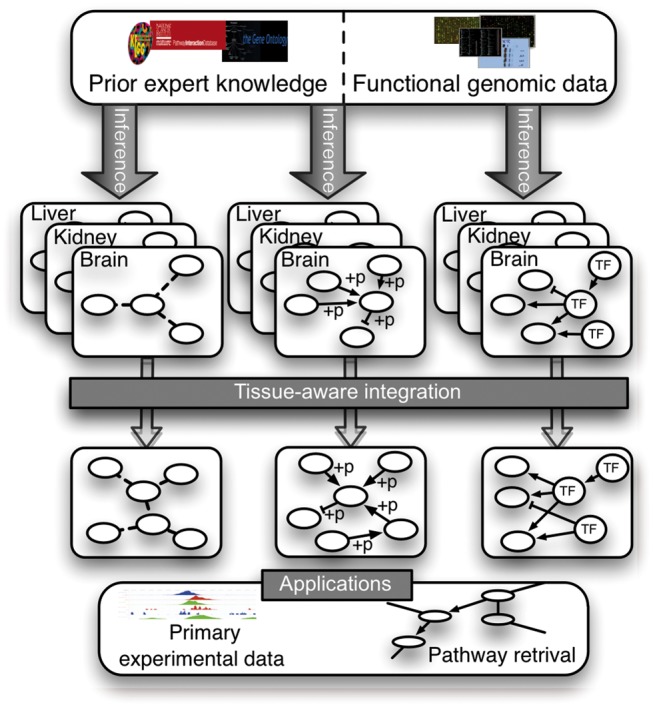
Schematic of our tissue-aware integrative pipeline for inferring metazoan gene interactions. We collect tissue and interaction-type specific gold standards, restricted to protein pairs that are both expressed in the tissue, based on curated databases and expression profiling of total 77 tissues. Next, we infer each interaction type network by integrating the genomic data compendium independently in the context of each tissue (i.e. with tissue-specific learning examples), resulting in multiple intermediate tissue-context networks per interaction-type. Following, we integrate the tissue-context based predictions to obtain the most probable set of labels for each gene pair across the interaction types. Finally, our predicted networks can be used in multiple applications such as pathway retrieval or aiding the interpretation of condition-specific primary datasets
