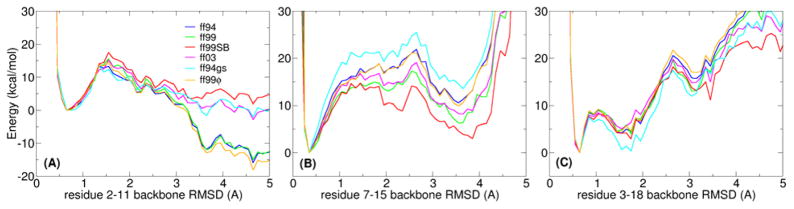
Figure 4.
“Lowest energy” profiles for three prototypical decoy systems, each tested with six AMBER force field variants (ff94, ff99, ff99SB, ff03, ff94gs, ff99ϕ): (A) trpzip2, which is known to adopt a β-hairpin, (B) a Baldwin-type sequence as a representative of an α-helix, and (C) trpcage as a representative of mixed secondary structures. RMSD values are calculated using the experimentally determined structure as a reference. Ideally, a force field should show lowest energies for the lowest RMSD values.