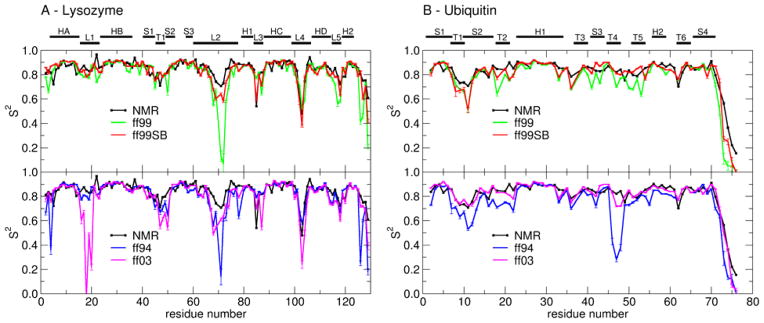
Figure 5.
Order parameters (S2) derived from experiment (black line) and calculated from MD simulations in explicit water with different force field parameter sets: ff99 (green), ff99SB (red), ff94 (blue) and ff03 (magenta). Error bars reflect the convergence of calculated S2 values. (A) Lysozyme. Secondary structures of lysozyme are labeled: helix A (HA: residues 4–15), loop 1 (L1: 16–23), helix B (HB: 24–36), strand 1 (S1: 41–45), turn 1 (T1: 46–49), strand 2 (S2: 50–53), strand 3 (S3: 58–60), long loop 2 (L2: 61–78), 310 helix 1 (H1: 80–84), loop 3 (L3: 85–89), helix C (HC: 89–99), loop 4 (L4: 100–107), helix D (HD: 108–115), loop 5 (L5: 116–119) and 310 helix 2 (H2: 120–124). (B) Ubiquitin. Secondary structure elements in ubiquitin: strand 1 (S1: residues 1–7), turn 1 (7–10), strand 2 (S2: 10–17), turn 2 (T2: 18–21), helix 1 (H1: 23–34), turn 3 (T3: 37–40), strand 3 (S3: 41–44), turn 4 (T4: 45–48), turn 5 (T5: 51–54), helix 2 (H2: 56–59), turn 6 (T6: 62–65) and strand 4 (S4: 66–71). Differences are described in the text.