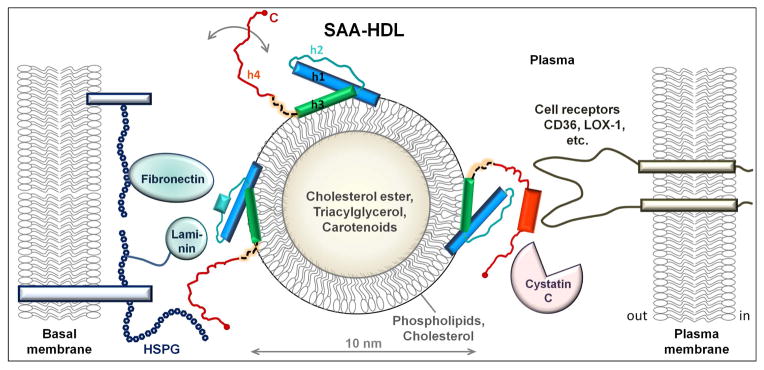
Figure 5.
Hypothetical model of SAA on HDL surface. SAA and HDL are drawn to scale. HDL is shown in cross-section to illustrate its polar surface containing amphipathic proteins, phospholipids (mainly phosphatidylcholine) and cholesterol, and the apolar core containing cholesterol esters, triacylglycerides and small amounts of fat-soluble vitamins such as carotenoids. Other HDL proteins such as apoA-I or apoA-II that are also present on SAA-HDL [25] are not shown. In the NT domain of SAA (residues 1–69 in blue, teal and green), amphipathic α-helices h1 and h3 form the putative HDL binding site. We posit that h1 and h3 on HDL retain their relative packing via the GPGG motif observed by x-ray crystallography. Parts of h1 and h3 that bind HDL are folded, accounting for an average α-helical content of ~35% observed in lipid-bound SAA by circular dichroism [25]. In the absence of additional ligands, the rest of the SAA molecule spans multiple conformations (circular arrow). The dynamic behavior of the CT domain (red) is facilitated by the flexible interdomain linker (dashed line). Apart from HDL, the NT domain can also bind other ligands, such as laminin and fibronectin that bind via h2 and h2/h3 linker [32, 33]. Alternatively, the NT domain of SAA can bind retinol via h3 (not shown) [14] and/or self-associate via h1 and h3 [14, 39]. CT domain can bind diverse ligands [64] including various cell receptors (CD36, LOX-1, etc.) [37], heparan sulfate proteoglycans (HSPG) [28], cystatin C [31], cholesterol ester hydrolase [58] and others. Ligand binding is probably augmented by the high intrinsic helical propensity of h4 (Fig. S2) that is expected to undergo coupled binding and folding. Thus, in the presence of additional bound ligands, the helical content in SAA on HDL may ultimately approach that seen in the crystal structure. SAA brings together ligands that are bound to NT- and CT-domains of the same SAA molecule, as well as those bound to different SAA molecules on the same HDL particle. Such bridging of diverse ligands, which may bind with positive or negative cooperativity depending on the environment, can modulate their functional interactions. We propose that SAA acts as a molecular hub modulating a wide range of functional interactions among diverse proteins, lipids, and proteoglycans.