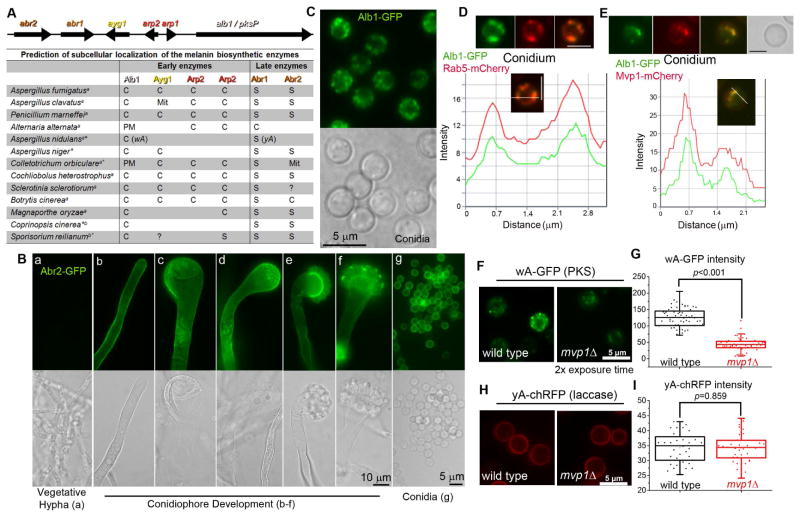
Figure 2. Distinct localization patterns for the early and late melanin enzymes.
(A) The melanin gene cluster in A. fumigatus and the predicted localization of melanin enzymes in diverse fungal species. S: Secreted protein; C: Cytosolic protein; PM: Plasma membrane; Mit: Mitochondria; a: Ascomycetes; b: Basidiomycetes; *: melanin genes not arranged in a cluster; ?: the presence of an ortholog is uncertain.
(B) The expression and localization of Abr2 during development. [a] vegetative hypha; [b] young stalk; [c–d] mature stalk; [e–f] young conidiophore; [g] conidia.
(C) Localization of Alb1 in conidia.
(D) Localization of Alb1-GFP and the endosomal marker Rab5-mCherry, and a fluorescence intensity plot along a cellular axis indicated with a white line.
(E) Localization of Alb1-GFP and Mvp1-mCherry, and a fluorescence intensity plot showing co-localization.
Localization(F) and fluorescence intensity (G) of the early melanin enzyme wA-GFP in WT and mvp1Δ in A. nidulans.
Localization (H) and fluorescence intensity (I) of the late melanin enzyme yA-chRFP in WT and mvp1Δ. Scale bars in panels D–F: 2.5 μm.
See also Figure S2