Figure 2. The AP2 transcription factor APTF-1 controls FLP-11 expression in RIS.
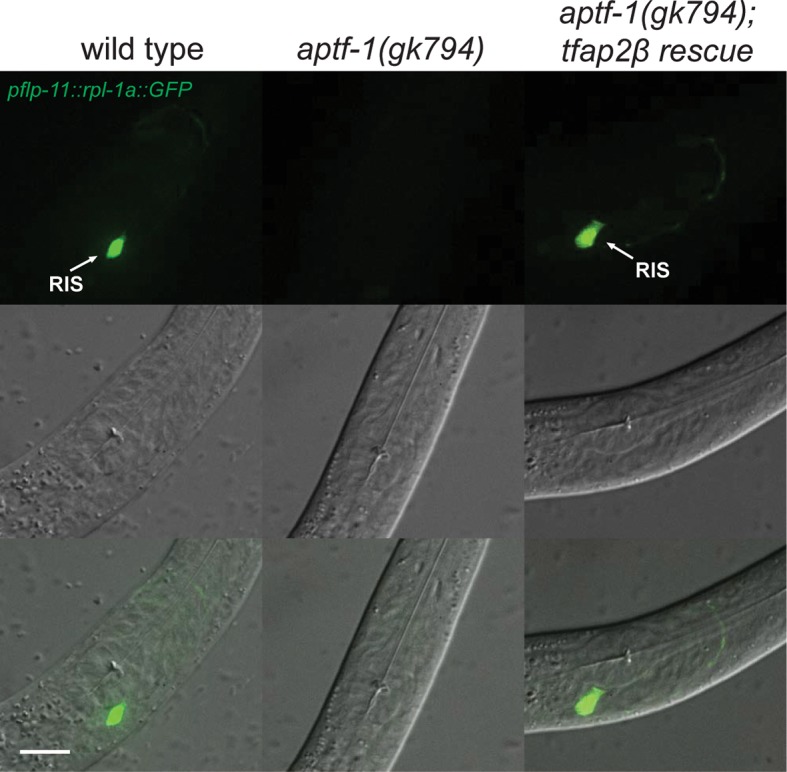
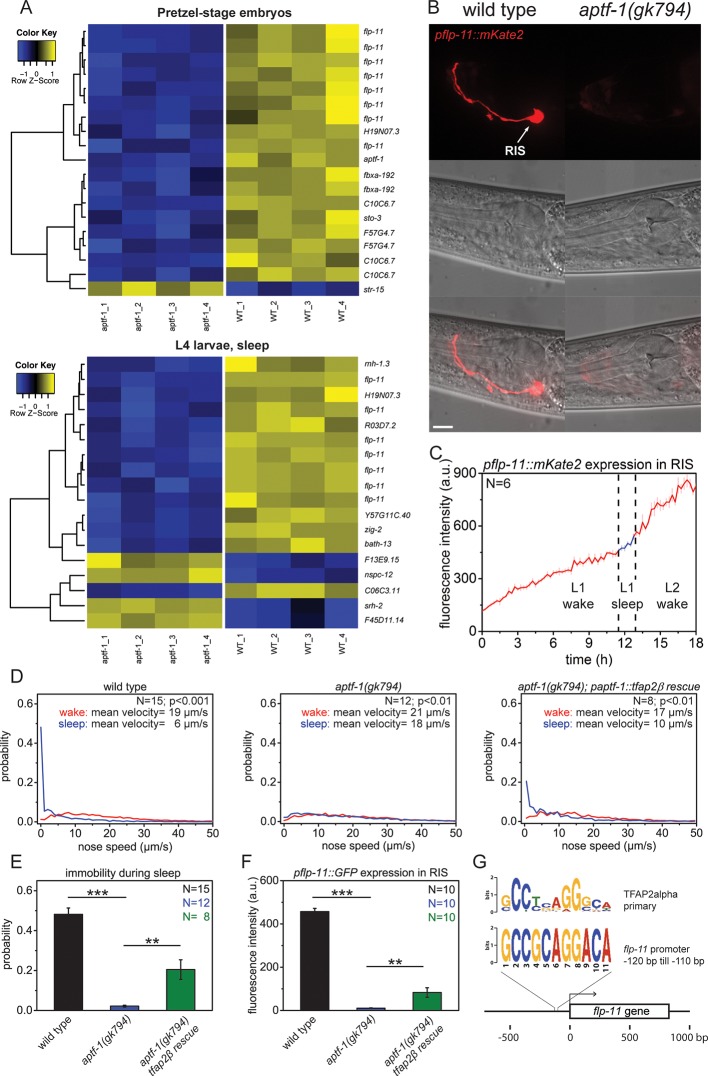
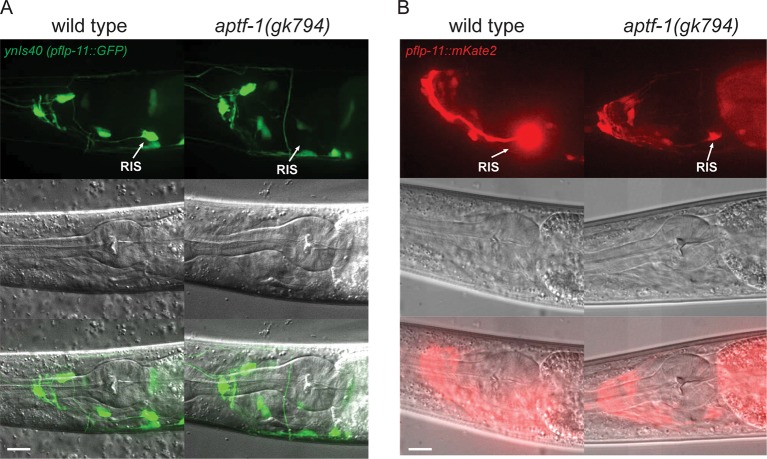
(A) Transcriptional analysis of aptf-1(gk794) mutants revealed genes that are regulated by APTF-1. Wild-type and aptf-1(gk794) pretzel-stage embryos and sleeping L4 larvae were used for a transcriptome analysis. In both life stages, expression of the FMRFamide-like neuropeptide FLP-11 was strongly reduced in aptf-1(gk794). This suggests transcriptional control of FLP-11 by APTF-1. Data can be found in Supplementary file 1, Tables 1A and 1B. (B) Expression of pflp-11::mKate2 in wild type and aptf-1(gk794). Expression of mKate2 was absent in RIS in aptf-1(gk794) showing that APTF-1 controls expression of FLP-11. Expression of flp-11 in RIS was reminiscent to the expression of the flp-11 homolog afp-6 in RIS in Ascaris nematodes (Yew et al., 2007). Expression for additional genes can be found in Figure 2—figure supplements 2 and 3. (C) flp-11 expression profile in RIS over the sleep-wake cycle. Expression does not change with the sleep-wake cycle. (D) Probability distribution of nose speeds during wake and sleep for wild type, aptf-1(gk794) and aptf-1(gk794); paptf-1::tfap2β rescue. (E) Comparison of immobility during sleep for wild type, aptf-1(gk794), and aptf-1(gk794); paptf-1::tfap2β. The mouse TFAP2β partially rescued the aptf-1(gk794) sleep phenotype. (F) Comparison of pflp-11::GFP fluorescence intensity in RIS for wild type, aptf-1(gk794), and aptf-1(gk794); paptf-1::tfap2β. The mouse TFAP2β partially rescued the expression of flp-11 in RIS (18% of wild-type level). (G) Analysis of putative AP2-binding sites in the flp-11 promoter region. The flp-11 promoter region was scanned for the primary mouse AP2α-binding site. Overlap was found (p<0.001, q=0.06 (Grant et al., 2011)) for one binding site. Statistical test used was Wilcoxon Signed Paired Ranks test. ** denotes statistical significance with p<0.01, *** denotes statistical significance with p<0.001. Scale bar is 10 µm.
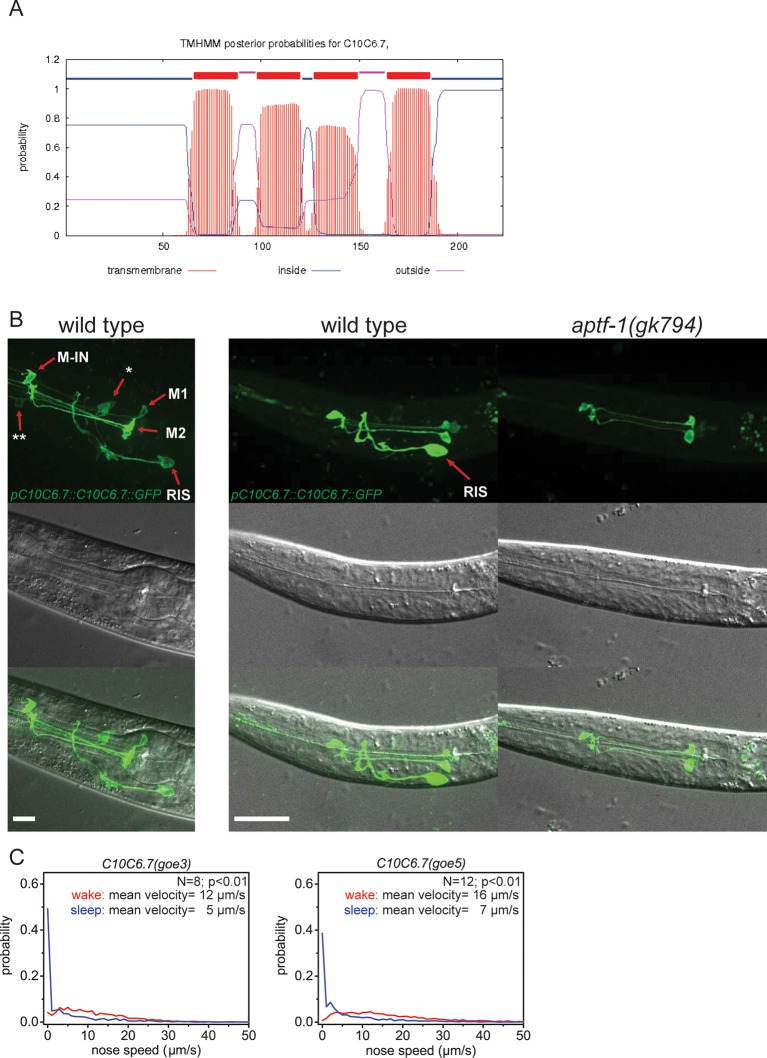
Figure 2—figure supplement 1. C10C6.7 is a putative four transmembrane helix protein that is expressed in RIS and that is controlled by aptf-1.
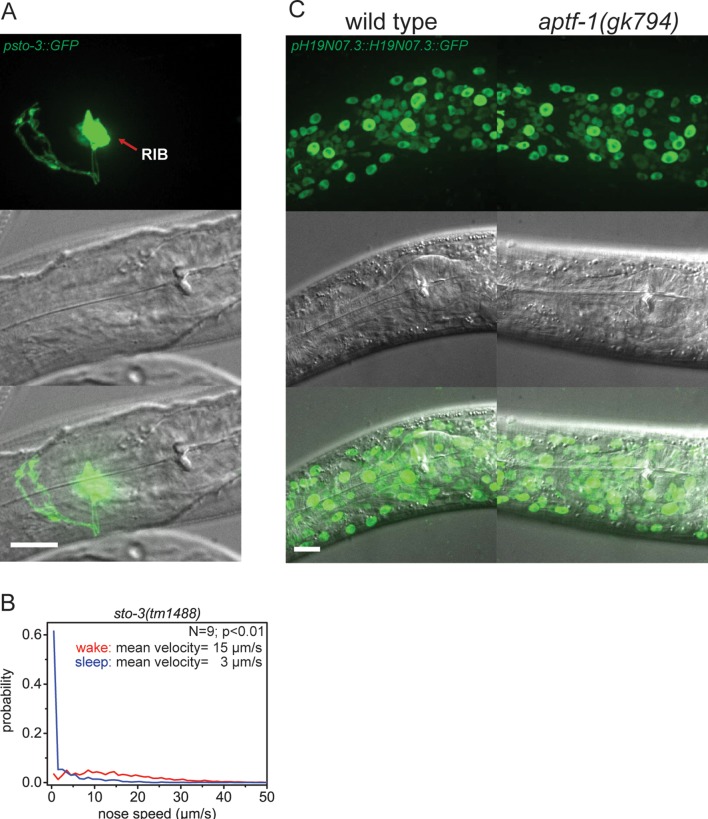
Figure 2—figure supplement 2. Expression pattern of sto-3 and H19N07.3.
Figure 2—figure supplement 3. FLP-11 is strongly expressed in RIS and weakly in additional neurons. APTF-1 controls the expression in RIS.
Figure 2—figure supplement 4. Mouse TFAP2beta partially restores expression of flp-11 neuropeptides in RIS in aptf-1 mutant worms.