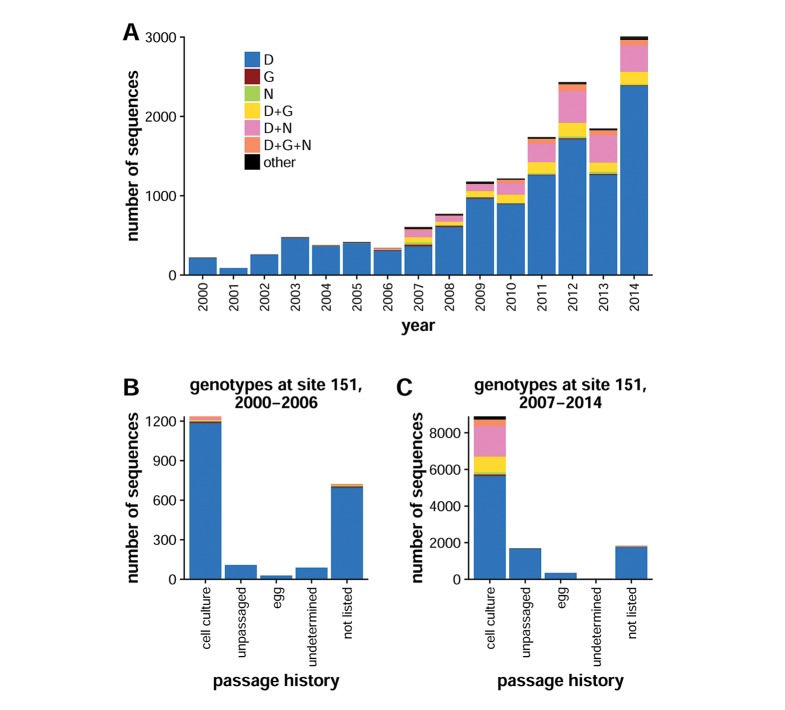
Figure 1. Ambiguous identities are common at NA site 151 after 2007.
(A) Shown are the number of human H3N2 influenza NA sequences in the GISAID EpiFlu database with the given identity at site 151 for each year from 2000 to 2014. Since 2007, ambiguous amino-acid identities have been present at residue 151 in about 20% of sequences. Sequences from (B) 2000 to 2006 and (C) 2007 to 2014 were classified into groups based on their passage history. Ambiguous amino-acid identities were present almost exclusively in isolates that had been passaged in cell culture. Sequences were classified as 'undetermined' if the passage history was difficult to interpret and as 'not listed' if the passage history was absent altogether. Mixed genotypes were inferred on the basis of IUPAC nucleotide ambiguity codes; for instance, the triplet GRT could refer to GAT or GGT, corresponding to amino acids D and G, respectively. Genotypes are indicated if they exceeded a frequency of 0.5% among all analyzed sequences; otherwise, they are categorized as 'other.' The computer code used for analysis is available in Figure 1—source data 1.
DOI: http://dx.doi.org/10.7554/eLife.13974.004