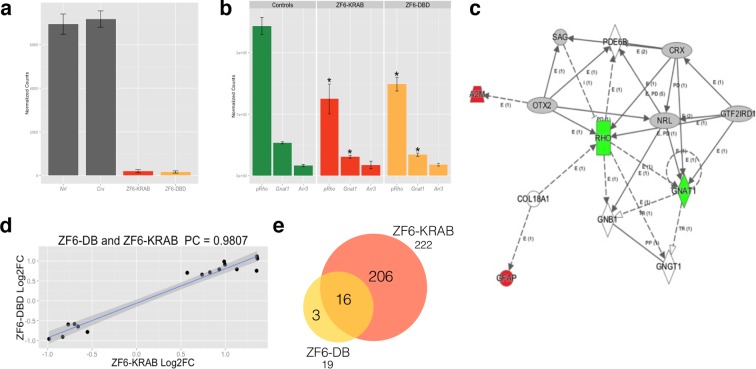
Figure 2. Photoreceptor delivery of ZF6-DB resulted in reduced genome-wide transcript perturbations.
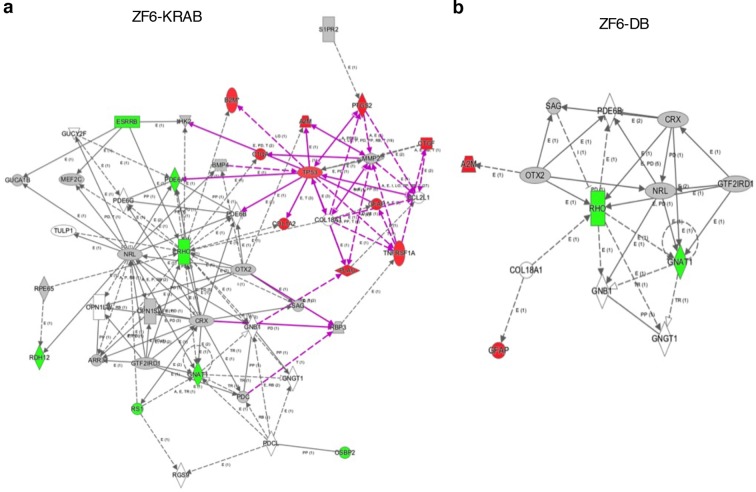
(a) RNA-Seq expression levels (Mean Normalized Counts) comparison between 2 endogenous TFs (Crx and Nrl) and the expression levels resulting from transduction of AAV8-CMV-ZF6-DB and AAV8-CMV-ZF6-KRAB, 15 days after retinal delivery (AAV8-CMV-ZF6-DB n= 6; AAV8-CMV-ZF6-KRAB n= 4 and 7 controls, non-transduced area). (b) Rho and rod Gnat1 and Cone Arrestin 3 expression levels in treated and control retina. (c) Ingenuity Pathway Analysis of DEGs after ZF6-DB AAV delivery in porcine retina showed a network of 13 genes. The 2 phototransduction genes RHO and GNAT1 are shown in green (down-regulated) whereas the 2 genes associated with primary inflammatory response network, A2M and GFAP, are up-regulated (red). (d) Transcriptional activation and repression concordances among Log Fold Changes of the genes in common (Swaroop et al., 2010) between ZF6-DB and ZF6-KRAB (Pearson Correlation Test; PC=0.9787; p value << 1x10-5). (e) Venn Diagrams, pairwise intersection of the 2 sets of Differentially Expressed Genes (DEGs). An adjusted p value (False Discovery Rate; FDR ≤ 0.1), without filtering on fold change levels, resulted in 19 and 222 DEGs, in ZF6-DBD and ZF6-KRAB treated retina, respectively. The intersection resulted significant by hypergeometric test (p value << 1x10-5).