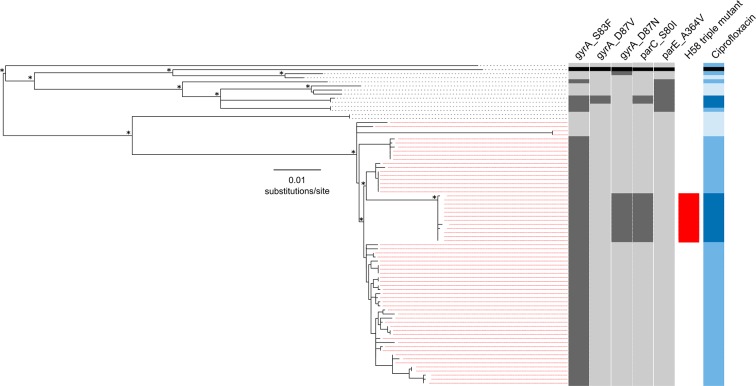
Figure 1. The phylogenetic structure of 78 Nepali Salmonella Typhi isolated during a gatifloxacin versus ceftriaxone randomised controlled trial.
Maximum likelihood phylogeny based on core-genome SNPs of 78 Salmonella Typhi RCT isolates with the corresponding metadata, including the presence of mutations (dark grey) in gyrA (S83F, D87V and D87N), parC (S80I) and parC (A364V) and susceptibility to ciprofloxacin (susceptible, light blue; intermediate, mid-blue and non-susceptible, dark blue) by Minimum Inhibitory Concentration (MIC). The reference strain CT18 was used for context and highlighted by the black boxes. Red lines linking to metadata show isolates belonging to the Salmonella Typhi H58 lineage (with H58 triple mutants highlighted), other lineages (non-H58) are shown with black lines. The scale bar indicates the number of substitutions per variable site (see methods). Asterisks indicate ≥85% bootstrap support at nodes of interest.