Abstract
Background
Silicosis is one of several severe occupational diseases for which effective diagnostic tools during early development are currently unavailable. In this study we focused on proteomic profiling during the early stages of silicosis to investigate the pathophysiology and identify the proteins involved.
Methods
Two-dimensional (2D) gel electrophoresis and MALDI-TOF-MS were used to assess the proteomic differences between healthy individuals (HI), dust-exposed workers without silicosis (DEW) and silicosis patients (SP). Proteins abundances that differed by a factor of two-fold or greater were subjected to more detailed analysis, and enzyme linked to immunosorbent assay (ELISA) was employed to correlate with protein expression data.
Results
Compared with HI, 42 proteins were more abundant and 8 were less abundant in DEW, and these were also differentially accumulated in SP. Closer inspection revealed that serine protease granzyme A, alpha-1-B-glycoprotein (A1BG) and the T4 surface glycoprotein precursor (TSGP) were among the up-regulated proteins in DEW and SP. Significant changes in serine proteases, glycoproteins and proto-oncogenes may be associated with the response to cytotoxicity and infectious pathogens by activation of T cells, positive regulation of extracellular matrix structural constituents and immune response, and fibroblast proliferation. Up-regulation of cytokines included TNFs, interferon beta precursor, interleukin 6, atypical chemokine receptor 2, TNFR13BV, and mutant IL-17F may be involved in the increased and persistent immune response and fibrosis that occurred during silicosis development.
Conclusions
Granzymes, glycoproteins, cytokines and immune factors were dramatically involved in the immune response, metabolism, signal regulation and fibrosis during the early development of silicosis. Proteomic profiling has expanded our understanding of the pathogenesis of silicosis, and identified a number of targets that may be potential biomarkers for early diagnosis of this debilitating disease.
Keywords: Cytokines, fibrosis, immune response, proteomic profiling, silicosis
Introduction
Silicosis is a severe disease affecting pulmonary function characterized by nodular lesions in the upper lobe of pulmonary system caused by inhalation of crystalline silica, a common mineral present in the earth’s crust, mines and many construction materials (1,2). These pulmonary lesions are irreversible without effective drugs following diagnosis by chest X-ray. Patients with silicosis are more susceptible to lung carcinoma and others severe diseases and silicosis is recognized as a severe occupational disease affecting public health and hygiene around the world. Reactive oxygen species (ROS), reactive nitrogen species (RNS), extracellular apoptosis related factor (ECM), cytokines and lipid-associated proteins were all reportedly involved in the development of silicosis (3,4). Unfortunately, an effective method involving biomarkers for early diagnosis remained unavailable at present.
Significant cellular and molecular alterations ensued following inhalation and deposition of silica minerals in lung tissues (5). Lower-level exposure to silica particles triggered reversible inflammatory lesions, whereas high-level exposure led to long-term effects on inflammation, cell proliferation, deposition of collagen and extracellular matrix components in mesenchymal cells (1). Alveolar macrophages (AM), neutrophils, T-lymphocytes and mast cells were all involved in fibrogenesis, and cross-talk between these cells played an important role in disease progression. In studies on the interaction of mast cells and neutrophils in mice, animals with intact mast cells endured more severe lung lesions than mast cell-deficient mice (6). Alveolar type I epithelial cells were involved in the early stages of fibrogenesis and may play an important role in repair and regeneration in this process (7). Up-regulation of early response proto-oncogenes c-fos, c-jun and c-myc may be critical to the initial stages of epithelial cell proliferation (8,9), and early response genes were associated with the development of apoptosis in epithelial cells. Silica inhalation can stimulate the cellular production of ROS which mediate the persistent inflammatory response associated with cell injury and fibrogenesis, and activated inflammatory cells may also release oxidants during silicosis (10). Both ROS and RNS were involved in the toxicity and signalling of fibrogenesis via effects on AM (11).
Growth factors and cytokines were notably affected in the granulomas of silicosis patients (SP), particularly transforming growth factor-beta (TGF-β) that played an important role in fibrogenesis and promoted the expression of collagen and fibronectin biosynthesis-related genes. Accumulation of epidermal growth factor (EGF) and transforming growth factor alpha (TGF-α) may play a crucial role in the mitogenesis of pulmonary epithelial cells and fibroblasts, and may stimulate these growth factor signaling pathways. Tumor necrosis factor (TNF) and interleukins were involved in the development of silicosis, and symptoms were alleviated with anti-TNF antibody or soluble TNF receptors under the experimental conditions (12). Up-regulated expression of TNF and IL-1 occurred prior to inflammation and fibrosis, and stimulates inflammatory and immune responses involving T- and B-cell lymphocyte proliferation and activation, and oxidative bursts (13,14). Research towards preventing and remedying fibrosis has focused on anti-oxidants, phospholipases and inhibiting the release of TNF and IL-1.
Treatment of silicosis is complex and difficult due to the complicated cross-talk that occurred between different signaling pathways and the impact that has on the patient’s condition. Studies on biomarkers for early diagnosis have focused on trace elements, ROS/RNS, superoxide dismutase (SOD), apoptosis-related factors, ECM, and cytokines, but specific and effective biomarkers for this disease remained elusive. In this study we focused on the proteomic profiling of SP to identify potential candidate proteins for early diagnosis. Serum samples were collected from healthy individuals (HI), dust-exposed workers (DEW) and diagnosed SP. Two-dimensional (2D)-gel electrophoresis and MALDI-TOF-MS were applied to identify differently accumulated proteins, and gene ontology (GO) classification using bioinformatics tools was subsequently performed. Enzyme linked to immunosorbent assay (ELISA) was then used for validation and quantitative analysis. These results improved our understanding of the mechanism of silicosis pathogenesis and provided several potential candidates for biomarkers that may be used for the early diagnosis of this disease.
Materials and methods
Samples collection
Three groups of whole blood samples were collected from HI, DEW and SP at the 8th People’s Hospital of Wuxi (Wuxi, China). Each group contained 15 individuals and their ages were from 55–64 years old (no significantly difference between three groups). For the selected individuals of HI, DEW and SP, each group was diagnosed exclusively and people who suffered from other diseases were excluded. Silicosis was diagnosed according to the Diagnostic criteria of pneumoconiosis of GBZ70-2009 (National Occupational Health Standards of the People’s Republic of China) by chest X-ray. DEW worked dust-exposed for 7–11 years but without remarkable symptom of silicosis. Permission for these studies was obtained from the medical ethics committee and all the subjects provided written informed consent using protocols that complied with the Declaration of Helsinki Principles. All blood samples were stored at 4 °C for 24 h, centrifuged at 4,000 rpm for 10 min (5418R, Eppendorf, Germany), and serum was stored at −80 °C for few days until used.
Protein extraction and removal of highly abundant proteins
In order to increase the reliability, five samples within groups were pooled into a new sample, consequently each group was performed in triplicate. Total proteins from HI, DEW and SP samples were extracted by using trichloroacetic acid, and Agilent human 14 multiple affinity removal system columns (Agilent, USA) were used removal of 14 high-abundance proteins (albumin, IgG, antitrypsin, IgA, transferrin, haptoglobin, fibrinogen, alpha2-macroglobulin, alpha1-acid glycoprotein, IgM, apolipoprotein AI, apolipoprotein AII, complement C3, and transthyretin) following the manufacturer’s instructions. Selective immunodepletion provided an enriched pool of low-abundance proteins for subsequent proteomic analysis and improved the resolution and dynamic range for 2D-gel electrophoresis and MS. Samples were subsequently dissolved in lysate solution (9 M urea, 4% CHAPS, 65 mM dithiothreitol and enzyme inhibitor cocktail) and quantified using Coomassie brilliant blue.
2d-gel electrophoresis
Each 120 µg sample in hydration solution (8 M urea, 2% CHAPS, 65 mM DTT, 0.50% IPG buffer) was loaded onto Immobiline Drystrip gels (18 cm, pH 3-10, GE Healthcare, USA) on an IPG-phor system (Amersham Bioscience, USA) for first dimensional electrophoresis. Isoelectric focusing (IEF) was conducted at 30 V for 6 h, 60 V for 6 h, 500 V for 1 h, 1,000 V for 1 h and 8,000 V for 20 h (total =64,000 Vhr). IEF gels were transferred to balanced solution (50 mM Tris HCl, 6 M urea, 2% SDS) and were vibrated for 15 min. Subsequently gels were sealed with low-melting point agarose, and SDS-PAGE was performed at 4 W for 45 min, followed by 15 W until the bromophenol blue reached the bottom, on an Ettan DALT SIX vertical system (Amersham Bioscience, Sweden). Each HD, DEW and SP sample was performed in triplicate. Following visualization by sliver nitrate staining, gels was scanned with an Artix Scan 1010 plus (Microtek, Taiwan) and analyzed by Image Master 2D platinum software (GE Healthcare, USA). Protein spots that were at least two-fold different between groups were quantified, filtered and the background was subtracted as described previously (Shanghai Yeslab Biotechnology, China).
MALDI-TOF-MS assay
Selected protein spots were excised, transferred to 96-well plates and fragmented by using an ultrasonic system for 5 min. Fragments were destained with acetonitrile, dried in vacuum for 5 min, and heated in a water bath with 10 mM DTT and 50 µL NH4HCO3 at 56 °C for 1 h. After 55 mM iodoacetamide and 25 mM NH4HCO3 were added at room temperature, samples were incubated in the dark for 45 min. Samples were washed in 25 mM NH4HCO3, 50% acetonitrile and finally 100% acetonitrile. Trypsin (Promega, USA) was diluted with 25 mM NH4HCO3, added and incubated on ice for 30 min. Then protein spots were covered with NH4HCO3 and incubated at 37 °C for 24 h. Hydrolysis was terminated with 0.1% trifluoroacetic acid (TFA).
Tryptic peptides were identified by MS, using an Autoflex II MALDI-TOF/MS (BRUKER, Germany) in a positive mode. Peptide mass fingerprint (PMF) maps were analyzed by Flexanalysis 3.0 with the SNAP algorithm and a signal-to-noise ratio of 1.5 (Bruker Daltonik, Germany). The Mascot search engine in Matrixscience (www.matrixscience.com) was used to generate PMF data that was subsequently searched against the NCBI redundant database with the trypsin cleavage parameter setting, a peptide mass tolerance of ±0.2 Da and a maximum of one missed cleavages (15).
ELISA assay
The expression levels of apolipoprotein, granzyme A and cathepsin-G were measured by using ELISA assay. Serum samples from DEW, SP and HI were added to the ELISA plates (Shanghai Yeslab Biotechnology, China) that were then closed with membranes and incubated for 30 min at 37 °C. Wells were washed five times with washing buffer, 50 µL HRP-conjugated reagents were added, and plates were incubated at 37 °C for 30 min. Following five more washes, 50 µL of chromogen solutions A and B were added and mixed at 37 °C for 15 min in the dark. After addition of stop solution, plates were assayed in a microplate reader (FC, Thermo Scientific, Finland) at 450 nm. A standard density curve was plotted, according to manufacturer’s protocol and samples density calculated by using the appropriate curve equation.
Data analysis
Differentially accumulated proteins identified in 2D-gel electrophoresis were log-transformed and hierarchical clustered using the centroid linkage method in Cluster V3.0 and visualized by using cluster grams in Java TreeView (V1.1.6r2) (16,17). Protein sequences were imported and searched against the non-redundant protein sequence database in CBI RefSeq using Blast2GO (V2.7.2) for gene-ontology annotations (18). Differentially accumulated proteins were analyzed by using t-test (SAS 9, USA).
Results
Protein expression profiling
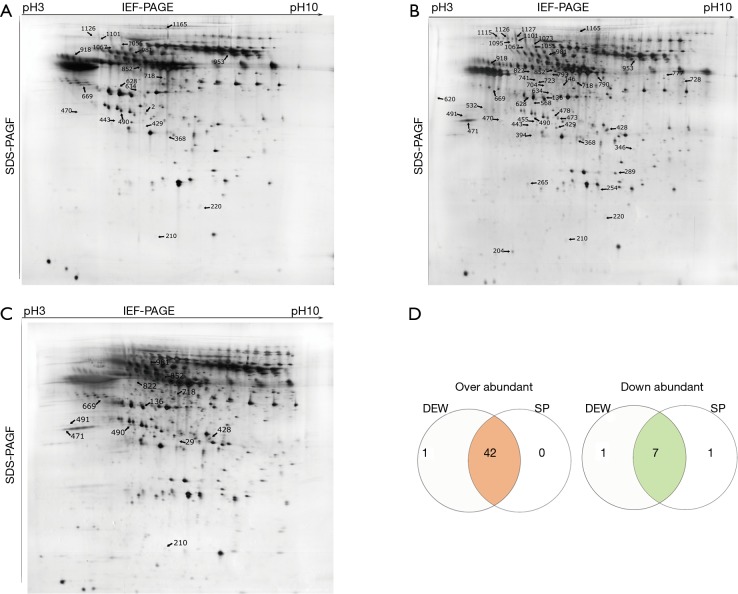
Serum samples from HI, DEW and SP were separated using 2D gels as described above, and a total of 313 differentially accumulated protein spots were detected that exhibited at least a two-fold difference between the HI, DEW and SP groups (Figure 1A-D). From the MALDI-TOF-MS assay, 51 unique proteins were successfully identified (Table 1).
Figure 1.
Two-dimensional gels of serum proteins from healthy individuals (HI), dust-exposed workers without silicosis (DEW) and diagnosed silicosis patients (SP) performed on IPG gel strips (17 cm, pH 3–10 L) followed by vertical SDS-PAGE. Identified proteins are indicated with arrows and code numbers. (A) Proteins upregulated in DEW compared to HI; (B) proteins upregulated in HI compared to DEW and SP; (C) proteins upregulated in SP compared to DEW; (D) venn diagram proteins up- or downregulated at least two-fold in DEW and SP compared with HI. The number of co-accumulated proteins in the two groups is shown in the intersections.
Table 1. Differentially expressed proteins identified by mass spectrometry from the serum of healthy individuals (HI), dust-exposed workers (DEW) and diagnosed silicosis patients (SP).
| Spot ID | Protein name | Genbank accession | Protein scores | Matched | Sequence coverage (%) | AA | DEW vs. HI | SP vs. HI |
|---|---|---|---|---|---|---|---|---|
| 428 | Chain B, cathepsin-G | 1KYN_B | 192 | 15 | 44 | 235 | Up | Up |
| 471 | Tumor necrosis factor receptor 13B variant | BAD97173 | 95 | 10 | 27 | 293 | Up | Up |
| 728 | Chain A, the crystal structure of the zymogen catalytic domain of complement protease C1r | 1GPZ_A | 92 | 11 | 25 | 399 | Up | Up |
| 532 | Chain A, crystal structure of P38 alpha in complex with Dp1376 | 3NNU_A | 182 | 14 | 42 | 354 | Up | Up |
| 1127 | Keratin, type II cuticular Hb4 | NP_149034 | 112 | 16 | 30 | 600 | Up | Up |
| 1115 | Furin preproprotein | NP_002560 | 185 | 19 | 22 | 794 | Up | Up |
| 1095 | ATP-dependent zinc metalloprotease YME1L1 isoform 3 | NP_055078 | 150 | 18 | 32 | 716 | Up | Up |
| 204 | Chain A, human growth hormone and extracellular domain of its receptor: crystal structure of the complex | 3HHR_A | 189 | 12 | 40 | 190 | Up | Up |
| 793 | Alpha-1-B-glycoprotein-human | OMHU1B | 126 | 15 | 29 | 474 | Up | Up |
| 704 | Chain A, 2.85 A crystal structure of PEDF | 1IMV_A | 135 | 13 | 25 | 398 | Up | Up |
| 346 | PTGER4 protein, partial | AAH27934 | 92 | 7 | 21 | 229 | Up | Up |
| 723 | IFNAR1 protein | AAH02590 | 121 | 10 | 32 | 387 | Up | Up |
| 254 | Chain A, human mannose binding protein carbohydrate recognition domain trimerizes through A triple alpha-helical coiled-coil | 1HUP_A | 76 | 7 | 43 | 141 | Up | Up |
| 1073 | X-linked interleukin-1 receptor accessory protein-like 2 | AF181285_1 | 122 | 16 | 25 | 658 | Up | Up |
| 790 | Chain B, crystal structure of human angiotensinogen complexed with renin | 2X0B_B | 121 | 19 | 31 | 452 | Up | Up |
| 478 | ICOS ligand isoform a precursor OR B, OR X1 | NP_056074 | 167 | 13 | 32 | 302 | Up | Up |
| 394 | Sex hormone-binding globulin isoform 6 | NP_001276044 | 136 | 11 | 60 | 235 | Up | Up |
| 146 | Chain A, thioredoxin peroxidase B from red blood cells | 1QMV_A | 126 | 11 | 47 | 197 | Up | Up |
| 265 | Tumor necrosis factor | AAA61198 | 147 | 12 | 32 | 233 | Up | Up |
| 568 | Proto-oncogene c-Fos | NP_005243 | 134 | 12 | 32 | 380 | Up | Up |
| 741 | Unnamed protein product | BAG60758 | 68 | 11 | 29 | 415 | Up | Up |
| 777 | Kininogen 1 variant, partial | BAD97309 | 185 | 20 | 46 | 427 | Up | Up |
| 473 | Ficolin-3 isoform 1 precursor | NP_003656 | 147 | 15 | 40 | 299 | Up | Up |
| 289 | Cytotoxic T-lymphocyte protein 4 isoform CTLA4-TM precursor | NP_005205 | 89 | 8 | 28 | 223 | Up | Up |
| 620 | src substrate cortactin isoform X2 | XP_006718511 | 67 | 12 | 34 | 369 | Up | Up |
| 455 | HLA-DRB1 protein | AAH31023 | 172 | 13 | 33 | 266 | Up | Up |
| 136 | Mutant IL-17F | ADY18335 | 70 | 5 | 39 | 163 | Up | Up |
| 822 | T4 surface glycoprotein precursor | AAA16069 | 76 | 9 | 23 | 458 | Up | Up |
| 491 | Unnamed protein product | CAA28465 | 144 | 15 | 42 | 321 | Up | Up |
| 29 | Tumor necrosis factor ligand superfamily member 4 isoform X1 | XP_005245532 | 227 | 15 | 59 | 133 | Up | Up |
| 210 | Interferon beta precursor | NP_002167 | 192 | 15 | 40 | 187 | Up | Up |
| 429 | Serum vitamin D-binding protein precursor | AAA52173 | 183 | 23 | 45 | 474 | Up | Up |
| 1126 | Coiled-coil Domain-containing protein 15 isoform X2 | XP_006718980 | 72 | 11 | 12 | 779 | Up | Up |
| 1067 | Unnamed protein product (fibulin-1 C) | BAG62463 | 80 | 14 | 17 | 641 | Up | Up |
| 953 | N-acetylmuramoyl-L-alanine amidase precursor | NP_443122 | 118 | 19 | 35 | 576 | Up | Up |
| 1055 | Complement component 9, Isoform CRA_b | EAW55988 | 125 | 19 | 32 | 567 | Up | Up |
| 1101 | Interleukin 12 receptor, beta 1, isoform CRA_c, partial | EAW84655 | 83 | 10 | 12 | 753 | Up | Up |
| 634 | Atypical chemokine receptor 2 | NP_001287 | 137 | 11 | 20 | 384 | Up | Up |
| 443 | Granzyme A | P12544 | 91 | 8 | 30 | 262 | Up | Up |
| 220 | Interleukin 6 | AFF18412 | 250 | 19 | 54 | 211 | Up | Up |
| 470 | Chain A, dimeric Apoa-Iv | 3S84_A | 93 | 12 | 34 | 273 | Up | Up |
| 368 | Medullasin | BAA00128 | 105 | 9 | 24 | 237 | Up | Up |
| 981 | Parathyroid hormone/parathyroid hormone-related peptide receptor isoform X4 | XP_005265401 | 152 | 14 | 23 | 562 | Up* | Down |
| 718 | Transforming growth factor beta-1 precursor | NP_000651 | 170 | 14 | 36 | 390 | Down | Down |
| 852 | Dipeptidyl peptidase 1 | P53634 | 122 | 11 | 23 | 463 | Down | Down |
| 918 | Chain A, crystallographic analysis of the human vitamin D binding protein | 1J78_A | 96 | 8 | 24 | 458 | Down | Down |
| 1165 | COL2A1, partial | CAA34683 | 140 | 18 | 26 | 1160 | Down | Down |
| 490 | Deoxyribonuclease II | ACN63522 | 161 | 12 | 21 | 344 | Down | Down* |
| 628 | Apolipoprotein A-IV precursor | AAA51748 | 107 | 12 | 28 | 376 | Down | Down |
| 669 | Differentially expressed in FDCP 6 homolog (mouse), isoform CRA_b | EAX03822 | 113 | 16 | 42 | 376 | Down | Down |
| 2 | Chain A, X-ray crystal structure of human transthyretin at room temperature | 3U2I_A | 150 | 9 | 91 | 117 | Down | Down |
A total of 42 and eight proteins were more or less abundant, respectively, in DEW compared with HI, which was also the case for samples from SP compared with HI (Table 1). Additionally, seven and six proteins were up and down regulated, respectively, in SP compared with DEW (Table 1). These differentially accumulated proteins were hierarchically clustered based on Pearson correlation and visualized by using TreeView software (16,17). All serum samples were clustered into three groups (HI, DEW and SP) in the top tree of the dendrogram, which implied that the results within groups have a high consistency (Figure 2). According to the tree structure, differentially expressed proteins were mainly clustered into two groups that included up- and down-regulated proteins mainly divided into immune related proteins, glycoproteins, oncogenes, lyases, metalloenzymes, lipoproteins, TNF and other functional proteins (Figure 2).
Figure 2.
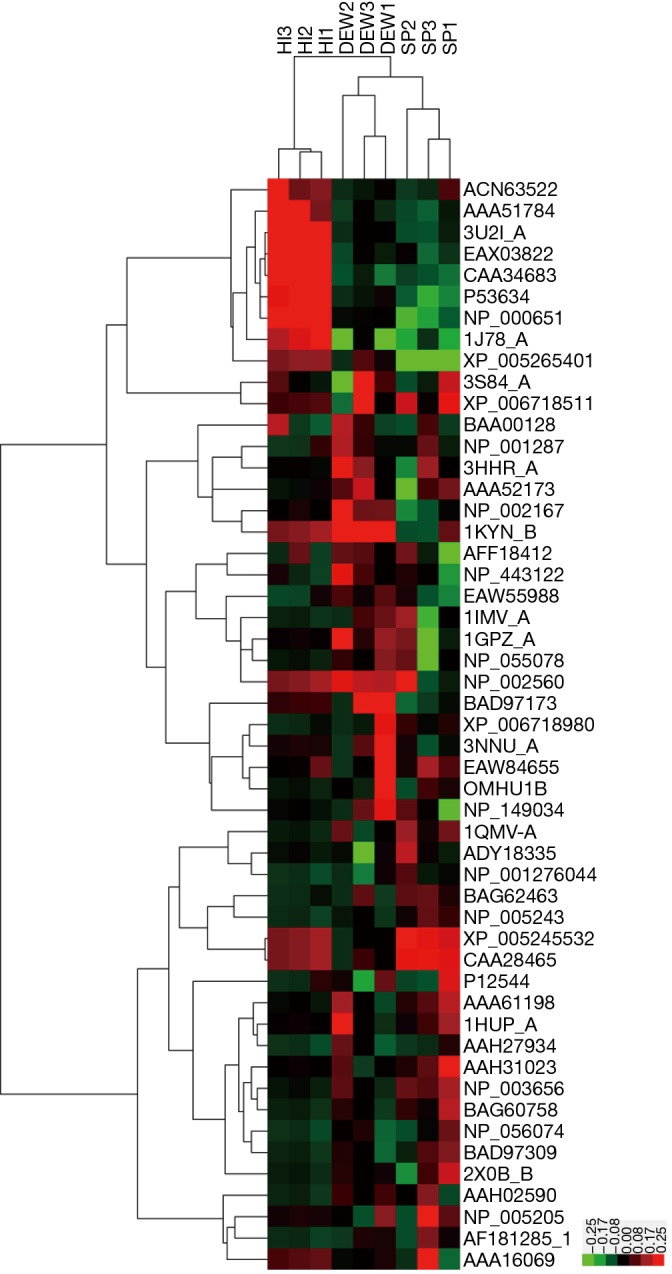
Heatmap of protein expression in HI, DEW and SP by hierarchical clustering analysis. All samples were arranged in columns and identified proteins were hierarchically clustered in centroid linkage in rows. Color blocks represent differentially expressed levels. Red: upregulated; green: downregulated.
GO enrichment analysis
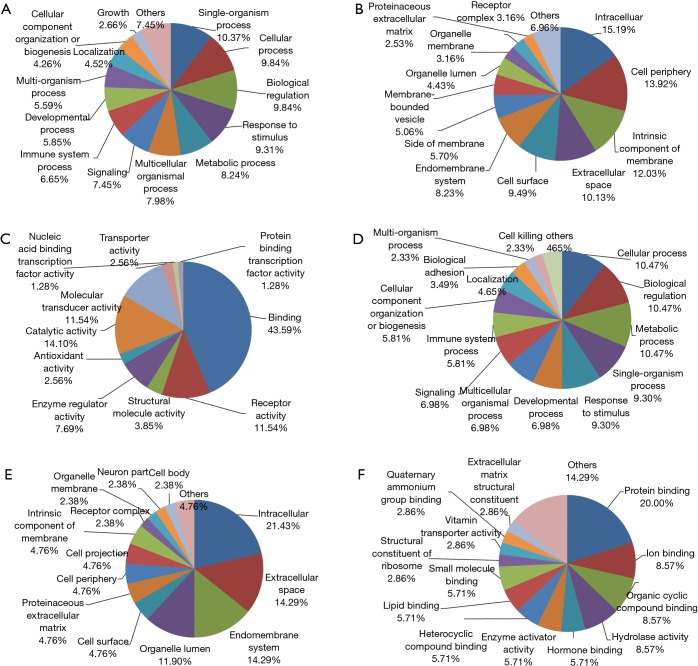
From the protein cluster analysis, identified proteins fell into up- and down-regulated categories (Figure 2), were annotated using the bioinformation tool (19). For proteins that were up-regulated in DEW and SP, GO classification enrichment resulted in three classifications of biological process, molecular function and cellular component. At the biological process level, the top six enriched terms were single-organism process, cellular process, biological regulation, response to stimulus, metabolic process and multicellular organismal process (Figure 3A). The cellular component class mainly consisted of intracellular, cell periphery, intrinsic component of membrane, extracellular space and cell surface categories, which accounted for over 50% of terms (Figure 3B). Protein binding, receptor activity, catalytic activity and molecular transducer activity accounted for 80% of the molecular function classification (Figure 3C).
Figure 3.
Differentially expressed proteins in HI, DEW and SP annotated by GO enrichment analysis. (A) GO enrichment of biological process classification of proteins upregulated in DEW and SP; (B) cellular component classification of proteins upregulated in DEW and SP; (C) molecular function classification of proteins upregulated in DEW and SP; (D) biological process classification of proteins upregulated in HI; (E) cellular component classification of proteins upregulated in HI; (F) molecular function classification of proteins upregulated in HI. HI, healthy individual; DEW, dust-exposed workers without silicosis; SP, silicosis patient; GO, gene ontology.
For proteins that were down-regulated in DEW, the top five enriched terms were cellular process, biological regulation, metabolic process, single-organism process and response to stimulus, which accounted for 50% of the biological process classification (Figure 3D). The cellular component class mainly consisted of intracellular, extracellular space, endomembrane system and organelle lumen, which accounted for more than half of this class (Figure 3E). At the molecular function level, the top four terms were associated with protein binding, ion binding, organic cyclic compound binding and hydrolase activity, which together accounted for 45% of this class (Figure 3F).
Validation of protein expression
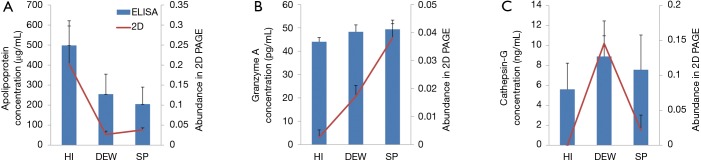
ELISA was employed to validate the expression differences of the identified proteins that included apolipoprotein, granzyme A and cathepsin-G, and the expression levels of these proteins were found to be highly consistent with the 2D-gel results (Figure 4). Compared with HI, apolipoprotein was down-regulated in DEW and SP groups, according to both 2D-gel and ELISA results (Figure 4A). By contrast, expression of granzyme A and cathepsin-G were up-regulated in DEW and SP in both methods (Figure 4B,C).
Figure 4.
Expression levels of candidate proteins analyzed by ELISA. The blue histogram bar represents protein concentration and expression levels are indicated on the left Y-axis. The red line represents protein abundance in 2D-PAGE and the scale is shown on the right Y-axis. The standard error of the mean is displayed for each protein. (A) Abundance of apolipoprotein; (B) abundance of granzyme A; (C) abundance of cathepsin-G. ELISA, enzyme linked immunosorbent assay.
Discussion
In this study we compared between the proteomic profiling of HI, DEW and SP, and found that the abundance of a number of cytokines, metabolism and extracellular matrix-related proteins, glycoproteins and granzyme-associated proteins were altered in the DEW and SP groups. These proteins were subsequently investigated in greater detail.
Cytokines
Cytokines are a broad group of small proteins or peptides that are released from immunocytes and that are involved in cell signaling and regulation of interactions in the development and differentiation of the immune response. We found that numerous cytokines including interleukins, interferons, chemokines, and TNFs were more abundant in DEW and SP compared with HI (Figure 2; Table 1). The TNF receptor superfamily member 13B (TNFRSF13B) was mainly expressed on the surface of B-cell subpopulations such as CD27+ memory B-cells, and bound to the TNF ligands B-cell activating factor and a proliferation-inducing ligand that functioned in the regulation of isotype switching, survival and B-lymphocyte differentiation in a complex functional network that was important for humoral responses (20,21). Coding variants of TNFRSF13B have been implicated in common variable immunodeficiency, and the variant TNFR13BV was found to be up-regulated in both DEW and SP, and the expression was two-fold higher in SP than in DEW (Table 1). This indicated that TNFR13BV may be involved in the increased and persistent immune response that occurred during silicosis development.
It was reported that TNFs, interleukins, interferons, transforming growth factors and chemokines were essential for pulmonary fibrosis, and persistent up-regulation of related cytokines activates fibroblasts, and caused fibrosis of the extracellular matrix (1,13,22,23). In this study TNF, TNF superfamily member 4 isoform X1, interferon beta precursor, IL-6, atypical chemokine receptor 2 and the mutant IL-17F were all up-regulated in DEW and SP (Figures 1,2, Table 1). Numerous inflammatory and immune responses including T-cell and B-cell proliferation and activation, and increased oxidative bursts and degranulation of inflammatory cells, were known to be stimulated by IL-1 and TNF. Chemokines played crucial roles in numerous physiological and pathological immune and inflammatory reactions, and atypical chemokine receptors were characterized by a lack of signaling and the ability to internalize and degrade chemokine ligands, which were required for regulation of inflammatory responses (24). These up-regulated proteins may be involved in the initiation of inflammation and subsequent signaling of fibroblasts.
The IL-1 receptor accessory protein was crucial for IL-33-induced activation of T-lymphocytes and mast cells, and IL1 receptor accessory protein-like was involved in X-linked mental retardation via interaction with neuronal calcium sensor-1 that regulated exocytosis. As a homologue of the IL-1 receptor accessory protein-like (IL1RAPL), IL1RAPL2 was responsible for X-linked mental retardation (MRX34) (25). We found that IL1RAPL2 was up-regulated in DEW and SP, indicating a role in the regulation of nervous signal transduction (Figures 1,2). Cytokine-associated proteins such as TNFs, interferon beta precursor, IL-6, atypical chemokine receptor 2 and mutant IL-17F were all highly abundant in DEW and SP, which may provoke crosstalk of the immune signalling involved in the increased and persistent immune response in silicosis development.
Extracellular matrix
Keratin or type II cuticular Hb4 (KRT4) is encoded by a member of the keratin gene family and is involved in the structural constituents of the cytoskeleton, regulation of keratinocyte differentiation, formation of keratin filaments, and hair follicle development. KRT4 was expressed at high levels in the blood of autoimmune of rheumatoid arthritis (RA) sufferers, and citrullination was detected in the synovial membranes of these patients (26). KRT4 was up-regulated in DEW and SP in this study (Table 1, Figure 1), but the function and possible regulatory role in silicosis required further investigation.
Human growth hormone hGH and the extracellular domain of its receptor (3HHR) were up-regulated in DEW and SP (Table 1; Figure 2). The binding of hGH to its receptor was crucial for regulating growth and development, and a sequential mechanism for dimerization mediated via binding domains may be important in signal transduction (27). 3HHR may be involved in the early stages of silicosis development through the positive regulation of the growth of multicellular organisms. The AK-STAT cascade was involved in growth hormone signaling pathways, and the JAK-STAT cascade in particular was involved in positive regulation of MAP kinase activity (Figure 3), by which cellular development and proliferation are regulated in the pulmonary system.
Glycoproteins
Glycoproteins contained oligosaccharide chains that were covalently attached to polypeptide side-chains following cotranslational or posttranslational modification. Secreted extracellular proteins were often glycosylated, and this mediates cellular interactions and signal transduction. Krebs von den lungen-6 (KL-6) was a glycoprotein belonging to the Mucin 1 (MUC1) family and was reported to be a sensitive marker for fibrosing lung diseases (28). KL-6 may be a chemotactic factor for fibroblasts and upregulation of KL-6 in the epithelial lining fluid in small airways may lead to intra-alveolar fibrosis in fibrosing pulmonary diseases (29). We found that alpha-1-B-glycoprotein (A1BG) and T4 surface glycoprotein precursor (TSGP) were both up-regulated in DEW and SP, and the expression was over two-fold higher in SP than DEW (Figure 1; Table 1). A1BG was reportedly involved in a number of diseases such as endometrial cancer, cervical cancer and cervical squamous cell carcinoma (30), and was also elevated in hepatic fibrosis patients, which allowed such patients to be easily distinguished from HI (31). TSGP was associated with T-cell costimulation, positive regulation of interleukin-2 biosynthesis, T-cell receptor complex formation, extracellular matrix structural constituents, the transmembrane receptor protein tyrosine kinase signaling pathway, early endosomes, innate immune responses, and proteinaceous extracellular matrix and MHC class II protein complexes. Up-regulation of both A1BG and TSGP in DEW indicated that these proteins may be involved in the early regulation of fibrosis in the lung and may be potentially useful for diagnosis factors.
Granzymes
Granzymes are serine proteases released by cytoplasmic granules within cytotoxic T-cells and natural killer cells that induced apoptosis within virus-infected cells. Cell death induced by granzyme A was mainly characterized by generation of single-stranded DNA nicks without the involvement of activated caspases. Granzyme A disrupts the mitochondrial inner membrane that subsequently releases ROS, and in response to ROS, the ER-associated SET complex translocates to the nucleus where granzyme A cleaves three SET complex subunits that are involved in DNA repair (32). Granzyme A may affect the nuclear lamins and histone H1 which are important for nuclear envelope stability and maintenance of chromatin structure that favor the activity of DNases (32). We found that granzyme A was up-regulated in DEW compared with HI (Figures 1,2, Table 1), which indicated that granzyme A may target cytotoxicity and clearance of infectious pathogens through activation of T-cells during the early developmental stages of silicosis. Similarly, granzyme B was up-regulated in infiltrating lymphocytes from patients with idiopathic pulmonary fibrosis, and in mononuclear cells following bleomycin treatment of wild-type mice (33).
Metabolism
Apolipoproteins transport lipids though lymphatic and circulatory systems, and regulate lipoprotein metabolism. We found that chain A of dimeric apolipoprotein A-IV (ApoA-IV) was up-regulated in DEW, however the precursor of this protein was down-regulated in DEW compared with HI (Table 1). This was consistent with a role in regulating the early development of silicosis. The ApoA-IV gene was linked to both ApoA-I and ApoC-III genes and located about 14 kb downstream of ApoA-I in the same orientation, indicating co-regulation of all the three genes (34). ApoA-IV participates in the regulation of various pathways including lipid absorption, transport and metabolism.
The ATP-dependent zinc metalloprotease YME1L1 isoform 3 was the human ortholog of yeast mitochondrial AAA metalloprotease, and was localized in mitochondria. YME1L was an integral membrane protein with the carboxy-terminus exposed to the inter-membrane space. Stable knockdown of YME1L caused impaired cell proliferation, apoptotic resistance, alterations to cristae morphology and diminished rotenone-sensitive respiration, and excessive accumulation of non-assembled respiratory chain subunits in the inner membrane (35). YME1L1 was up-regulated in both DEW and SP in this study (Table 1), indicating a role in the proteolytic regulation of respiratory chain biogenesis during initiation of silicosis.
Proto-oncogene related proteins
C-fos was firstly reported in rat fibroblasts as the transforming gene in Finkel-Biskis-Jinkins murine osteogenic sarcoma virus, was a proto-oncogene member of the Fos transcription factor family, and maps to chromosome region 14q21→q31. The c-fos protein forms a heterodimer with c-jun that results in the formation of the activator protein-1 (AP-1) complex which binds DNA at AP-1 specific sites, stimulating transcription of AP-1 responsive genes by converting extracellular signals (36). The FOS proteins appear to regulate cell proliferation, apoptotic cell death, differentiation and transformation. We found that the proto-oncogene c-fos was up-regulated in DEW and SP (Table 1; Figures 1,2). The inception of proliferation in fibroblasts by silica minerals may emerge after up-regulation of the early response proto-oncogenes c-fos and c-jun (37). The AP-1 complex binds promoter regions of target genes that governed inflammation, proliferation and apoptosis, and increased expression of early response genes may played an important role in the pathogenesis of pulmonary fibrosis.
Proteases
The serine protease cathepsin G is an important azurophilic granule protein in neutrophils and is crucial in the role of neutrophils in inflammation, modulation of integrin clustering, degradation of extracellular matrix components and cytokines, and direct chemo-attraction of leukocytes. In the present study, we found that chain B of cathepsin-G was up-regulated in DEW compared to HI (Figures 1,2; Table 1), but was down-regulated in SP compared with DEW (Table 1). Cathepsin-G may therefore be involved specifically in inflammatory processes that occurred during the early stages of fibrosis, and this protein may be a potential biomarker of early diagnosis of silicosis.
Furin is member of the proprotein convertase family characterized by subtilisin-related serine proteases, and is involved in the secretory and endocytic pathways in which it cleaves proproteins at clusters of basic residues. Multiple physiological substrates were known including growth factors, coagulation proteins, extracellular matrix components and protease precursors such as matrix metalloproteases (MMPs) that may contribute to both acute and chronic silicosis; an imbalance of MMPs and TIMPs may result in extracellular matrix remodeling and basement membrane disruption in silicosis (38). The numerous roles of furin in human pathophysiology have attracted interest in the potential targeting of this protein with therapeutic agents. The furin preproprotein was found to be up-regulated in both DEW and SP groups (Table 1; Figure 2), indicating a potential role in the early development of silicosis, and may therefore be a useful biomarker.
Other functions
The membrane-anchored HLA-DRB1 belongs to the HLA class II beta chain paralogues, and plays a crucial role in the immune system by presenting peptides derived from extracellular proteins. HLA-DRB1 alleles confer high-affinity binding to asparaginase epitopes, resulting in enhanced hypersensitivity (39). The expression of HLA-DR by AM was higher than lung and blood T-cells in sarcoidosis (40). We found that HLA-DRB1 was highly abundant in DEW and SP groups (Figures 1,2; Table 1), and may therefore be involved in initiating the immune response in the development of silicosis. Further study of HLA-DRB1 was needed to understand the potential involvement in silicosis pathogenicity.
Conclusions
In summary, we identified a number of proteins involved in silicosis, including granzyme A, a serine protease released from cytoplasmic granules. Granzyme A was up-regulated in DEW, indicating a contribution to cytotoxicity and clearance of infectious pathogens by activation of T-cells in the early development of silicosis. The high abundance of A1BG and TSGP in DEW and SP suggested these proteins may be involved in the positive regulation of extracellular matrix structural constituents and the immune response. Initiation of fibroblast activation by silica minerals may occur after up-regulation of early response proto-oncogenes, such as c-fos, which was also up-regulated in DEW and SP, as the proteases cathepsin G and furin that may be involved in inflammation in the early stages of fibrosis. These proteases were therefore potential biomarkers that may be useful for the early diagnosis of silicosis. Also up-regulated cytokines such as TNFs, interferon beta precursor, interleukin 6, atypical chemokine receptor 2, TNFR13BV, and mutant IL-17F that may be involved in the increased and persistent immune response in silicosis development. Together, these results showed that a large number of proteins and peptides were dramatically altered during the early development of silicosis. Additionally, proteomic profiling may assist our understanding of this debilitating disease and facilitate the discovery of potential biomarkers for early diagnosis.
Acknowledgements
Funding: This work was supported by the Jiangsu Provincial Special Foundation for People’s Livelihoods of Science and Technology (BL2014024) and the Jiangsu Provincial Foundation for Six Talent Summit (2014WSN063).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Mossman BT, Churg A. Mechanisms in the pathogenesis of asbestosis and silicosis. Am J Respir Crit Care Med 1998;157:1666-80. 10.1164/ajrccm.157.5.9707141 [DOI] [PubMed] [Google Scholar]
- 2.Castranova V, Porter D, Millecchia L, et al. Effect of inhaled crystalline silica in a rat model: time course of pulmonary reactions. Mol Cell Biochem 2002;234-235:177-84. 10.1023/A:1015967017103 [DOI] [PubMed] [Google Scholar]
- 3.Fubini B, Hubbard A. Reactive oxygen species (ROS) and reactive nitrogen species (RNS) generation by silica in inflammation and fibrosis. Free Radic Biol Med 2003;34:1507-16. 10.1016/S0891-5849(03)00149-7 [DOI] [PubMed] [Google Scholar]
- 4.Hata J, Aoki K, Mitsuhashi H, et al. Change in location of cytokine-induced neutrophil chemoattractants (CINCs) in pulmonary silicosis. Exp Mol Pathol 2003;75:68-73. 10.1016/S0014-4800(03)00029-7 [DOI] [PubMed] [Google Scholar]
- 5.Diseases associated with exposure to silica and nonfibrous silicate minerals. Silicosis and Silicate Disease Committee. Arch Pathol Lab Med 1988;112:673-720. [PubMed] [Google Scholar]
- 6.Suzuki N, Horiuchi T, Ohta K, et al. Mast cells are essential for the full development of silica-induced pulmonary inflammation: a study with mast cell-deficient mice. Am J Respir Cell Mol Biol 1993;9:475-83. 10.1165/ajrcmb/9.5.475 [DOI] [PubMed] [Google Scholar]
- 7.Lesur O, Bouhadiba T, Melloni B, et al. Alterations of surfactant lipid turnover in silicosis: evidence of a role for surfactant-associated protein A (SP-A). Int J Exp Pathol 1995;76:287-98. [PMC free article] [PubMed] [Google Scholar]
- 8.Heintz NH, Janssen YM, Mossman BT. Persistent induction of c-fos and c-jun expression by asbestos. Proc Natl Acad Sci U S A 1993;90:3299-303. 10.1073/pnas.90.8.3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janssen YM, Barchowsky A, Treadwell M, et al. Asbestos induces nuclear factor kappa B (NF-kappa B) DNA-binding activity and NF-kappa B-dependent gene expression in tracheal epithelial cells. Proc Natl Acad Sci U S A 1995;92:8458-62. 10.1073/pnas.92.18.8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rom WN, Bitterman PB, Rennard SI, et al. Characterization of the lower respiratory tract inflammation of nonsmoking individuals with interstitial lung disease associated with chronic inhalation of inorganic dusts. Am Rev Respir Dis 1987;136:1429-34. 10.1164/ajrccm/136.6.1429 [DOI] [PubMed] [Google Scholar]
- 11.Blackford JA, Jr, Antonini JM, Castranova V, et al. Intratracheal instillation of silica up-regulates inducible nitric oxide synthase gene expression and increases nitric oxide production in alveolar macrophages and neutrophils. Am J Respir Cell Mol Biol 1994;11:426-31. 10.1165/ajrcmb.11.4.7522485 [DOI] [PubMed] [Google Scholar]
- 12.Piguet PF, Vesin C. Treatment by human recombinant soluble TNF receptor of pulmonary fibrosis induced by bleomycin or silica in mice. Eur Respir J 1994;7:515-8. 10.1183/09031936.94.07030515 [DOI] [PubMed] [Google Scholar]
- 13.Driscoll KE, Hassenbein DG, Carter JM, et al. TNF alpha and increased chemokine expression in rat lung after particle exposure. Toxicol Lett 1995;82-83:483-9. 10.1016/0378-4274(95)03578-8 [DOI] [PubMed] [Google Scholar]
- 14.Driscoll KE, editor. In vitro evaluation of mineral cytotoxicity and inflammatory activity. G. D. Guthrie JaBTM, editor. Washington, DC: Mineralogical Society of America, 1993. [Google Scholar]
- 15.Perkins DN, Pappin DJ, Creasy DM, et al. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999;20:3551-67. [DOI] [PubMed] [Google Scholar]
- 16.de Hoon MJ, Imoto S, Nolan J, et al. Open source clustering software. Bioinformatics 2004;20:1453-4. 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- 17.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 1998;95:14863-8. 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conesa A, Götz S, García-Gómez JM, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005;21:3674-6. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 19.Götz S, García-Gómez JM, Terol J, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 2008;36:3420-35. 10.1093/nar/gkn176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng LG, Sutherland AP, Newton R, et al. B cell-activating factor belonging to the TNF family (BAFF)-R is the principal BAFF receptor facilitating BAFF costimulation of circulating T and B cells. J Immunol 2004;173:807-17. 10.4049/jimmunol.173.2.807 [DOI] [PubMed] [Google Scholar]
- 21.Sazzini M, Zuntini R, Farjadian S, et al. An evolutionary approach to the medical implications of the tumor necrosis factor receptor superfamily member 13B (TNFRSF13B) gene. Genes Immun 2009;10:566-78. 10.1038/gene.2009.43 [DOI] [PubMed] [Google Scholar]
- 22.Perdue TD, Brody AR. Distribution of transforming growth factor-beta 1, fibronectin, and smooth muscle actin in asbestos-induced pulmonary fibrosis in rats. J Histochem Cytochem 1994;42:1061-70. 10.1177/42.8.8027525 [DOI] [PubMed] [Google Scholar]
- 23.Matsuzaki H, Maeda M, Lee S, et al. Asbestos-induced cellular and molecular alteration of immunocompetent cells and their relationship with chronic inflammation and carcinogenesis. J Biomed Biotechnol 2012;2012:492608. [DOI] [PMC free article] [PubMed]
- 24.Graham GJ. D6 and the atypical chemokine receptor family: novel regulators of immune and inflammatory processes. Eur J Immunol 2009;39:342-51. 10.1002/eji.200838858 [DOI] [PubMed] [Google Scholar]
- 25.Ferrante MI, Ghiani M, Bulfone A, et al. IL1RAPL2 maps to Xq22 and is specifically expressed in the central nervous system. Gene 2001;275:217-21. 10.1016/S0378-1119(01)00659-X [DOI] [PubMed] [Google Scholar]
- 26.Chang X, Zhao Y, Wang Y, et al. Screening citrullinated proteins in synovial tissues of rheumatoid arthritis using 2-dimensional western blotting. J Rheumatol 2013;40:219-27. 10.3899/jrheum.120751 [DOI] [PubMed] [Google Scholar]
- 27.de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science 1992;255:306-12. 10.1126/science.1549776 [DOI] [PubMed] [Google Scholar]
- 28.Kohno N. Serum marker KL-6/MUC1 for the diagnosis and management of interstitial pneumonitis. J Med Invest 1999;46:151-8. [PubMed] [Google Scholar]
- 29.Hirasawa Y, Kohno N, Yokoyama A, et al. KL-6, a human MUC1 mucin, is chemotactic for human fibroblasts. Am J Respir Cell Mol Biol 1997;17:501-7. 10.1165/ajrcmb.17.4.2253 [DOI] [PubMed] [Google Scholar]
- 30.Canales NA, Marina VM, Castro JS, et al. A1BG and C3 are overexpressed in patients with cervical intraepithelial neoplasia III. Oncol Lett 2014;8:939-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin S, Zhou Y, Lok AS, et al. SRM targeted proteomics in search for biomarkers of HCV-induced progression of fibrosis to cirrhosis in HALT-C patients. Proteomics 2012;12:1244-52. 10.1002/pmic.201100601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieberman J, Fan Z. Nuclear war: the granzyme A-bomb. Curr Opin Immunol 2003;15:553-9. 10.1016/S0952-7915(03)00108-0 [DOI] [PubMed] [Google Scholar]
- 33.Miyazaki H, Kuwano K, Yoshida K, et al. The perforin mediated apoptotic pathway in lung injury and fibrosis. J Clin Pathol 2004;57:1292-8. 10.1136/jcp.2003.015495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elshourbagy NA, Walker DW, Boguski MS, et al. The nucleotide and derived amino acid sequence of human apolipoprotein A-IV mRNA and the close linkage of its gene to the genes of apolipoproteins A-I and C-III. J Biol Chem 1986;261:1998-2002. [PubMed] [Google Scholar]
- 35.Stiburek L, Cesnekova J, Kostkova O, et al. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol Biol Cell 2012;23:1010-23. 10.1091/mbc.E11-08-0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu R, Boyle WJ, Meek J, et al. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell 1988;54:541-52. 10.1016/0092-8674(88)90076-1 [DOI] [PubMed] [Google Scholar]
- 37.Janssen YM, Heintz NH, Marsh JP, et al. Induction of c-fos and c-jun proto-oncogenes in target cells of the lung and pleura by carcinogenic fibers. Am J Respir Cell Mol Biol 1994;11:522-30. 10.1165/ajrcmb.11.5.7946382 [DOI] [PubMed] [Google Scholar]
- 38.Pérez-Ramos J, de Lourdes Segura-Valdez M, Vanda B, et al. Matrix metalloproteinases 2, 9, and 13, and tissue inhibitors of metalloproteinases 1 and 2 in experimental lung silicosis. Am J Respir Crit Care Med 1999;160:1274-82. 10.1164/ajrccm.160.4.9808006 [DOI] [PubMed] [Google Scholar]
- 39.Fernandez CA, Smith C, Yang W, et al. HLA-DRB1*07:01 is associated with a higher risk of asparaginase allergies. Blood 2014;124:1266-76. 10.1182/blood-2014-03-563742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tapia FJ, Goihman-Yahr M, Cáceres-Dittmar G, et al. Leukocyte immunophenotypes in bronchoalveolar lavage fluid and peripheral blood of paracoccidioidomycosis, sarcoidosis and silicosis. Histol Histopathol 1991;6:395-402. [PubMed] [Google Scholar]