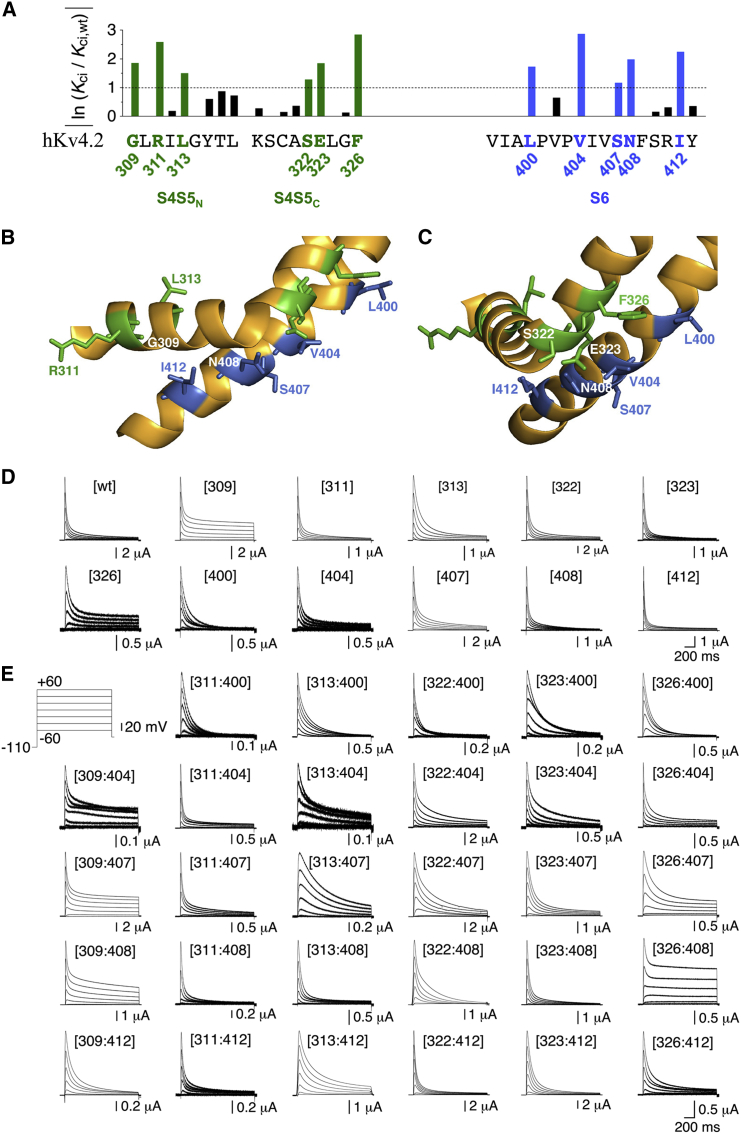
Figure 1.
Mutational analysis of S4S5/S6 dynamic binding in Kv4.2 channel CSI with monomeric constructs. (A) Effects of mutating individual S4S5 (left) and S6 (right) residues in Kv4.2 to alanine. The effects refer to low-voltage inactivation as a measure of CSI and are quantified as ln (Kci/Kci,wt) magnitude (Kci refers to a single closed-inactivated equilibrium constant (data based on Barghaan and Bähring (16); see Materials and Methods). The human Kv4.2 S4S5 and S6 native amino acid sequences in single letter code represent the x axis of the bar graph. S4S5 is subdivided in N-terminal (S4S5N, S4-S5 linker helix) and C-terminal (S4S5C, initial S5 segment) portions. An ln (Kci/Kci,wt) magnitude of >1 (dotted line) was arbitrarily chosen as a cutoff value for a significant effect (16). Significant effects on CSI when mutated to alanine were previously obtained for G309, R311, L313, S322, E323, and F326 in S4S5 (green), and for L400, V404, S407, N408, and I412 in S6 (blue) (16). (B and C) Structure of the S4S5/S6 interface in an individual α-subunit. Kv4.2 S4S5 and S6 homology modeling is based on the crystal structure of the Kv1.2-2.1 paddle chimera (19) (green, S4S5 residues; blue, S6 residues). In (B) the S4S5N (harboring G309, R311, and L313) is shown from the side. In (C) the S4S5/S6 interface is rotated by 90°, showing S4S5N from the front and offering a better view of the S4S5C residues S322, E323, and F326. In S6 the residues L400, V404, S407, N408, and I412 are shown. (D) Current families obtained under two-electrode voltage clamp from oocytes expressing monomeric Kv4.2 wt, S4S5 single mutants, and S6 single mutants. (E) Currents mediated by monomeric Kv4.2 S4S5/S6 double mutants (inset: voltage protocol; only test pulses between −60 and +60 mV in 20 mV increments are shown).