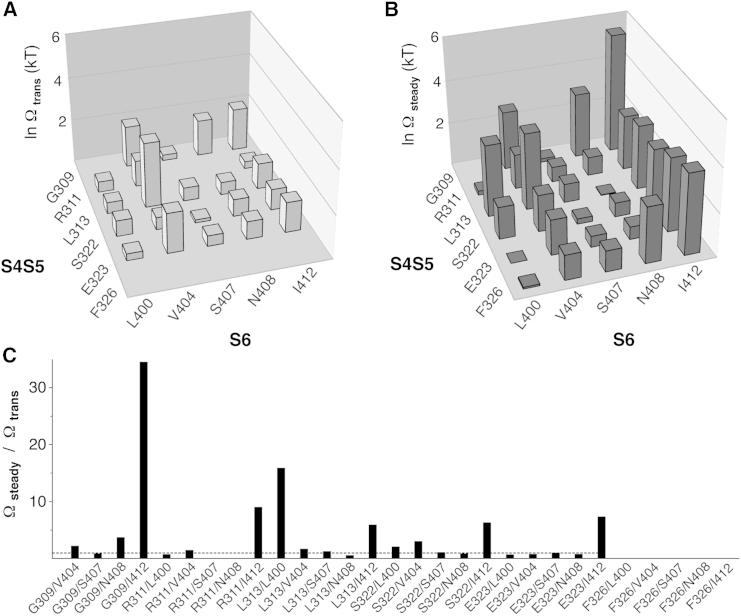
Figure 6.
Functional-coupling analysis for S4S5/S6 amino acid pairs based on monomeric Kv4.2 constructs. (A and B) Coupling coefficients for all S4S5/S6 pairings studied were determined by double-mutant cycle analysis based on Ktrans and Ksteady values and plotted as lnΩtrans (A) and lnΩsteady (B), respectively (see Materials and Methods). The three-dimensional plots in (A) and (B) show the native S4S5 amino acid residues on the left x axis and the native S6 amino acid residues on the right x axis. The mean lnΩtrans and lnΩsteady values are shown as vertical cuboids (common y axis, error bars omitted). Note the homogeneous and strong functional coupling during steady-state inactivation for pairings with the S6 residues V404 and I412. (C) The ratio Ωsteady/Ωtrans shows how functional coupling may change when the channel goes from transitory to steady-state inactivation (dotted line: Ωsteady/Ωtrans = 1, no change).