Figure 1. Overview of G. hansenii ATCC 53582 genome.
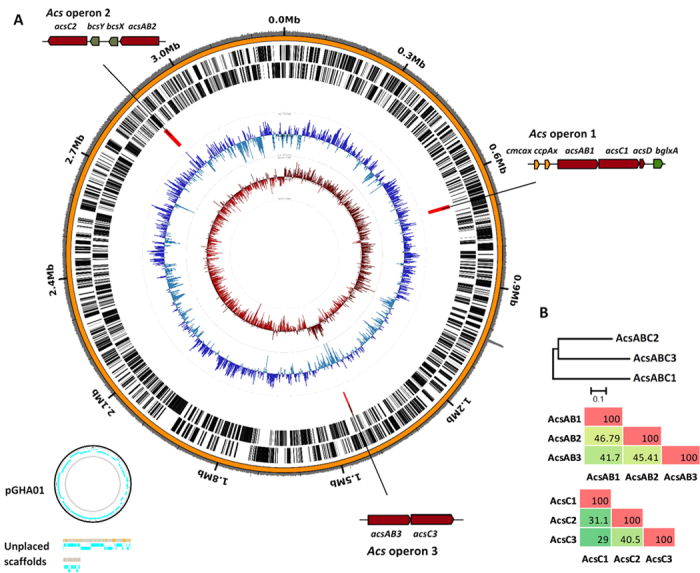
(A) G. hansenii ATCC 53582 genome is 3.27 Mbp in size, with a GC% of 59.48, and contains a predicted 2988 protein coding genes, 49 tRNAs, 1 tmRNA, and 7 rRNA genes. The genome contains a chromosome of approximately 3.27 Mbp and at least one 51 kb plasmid pGHA01. Additionally, it contains 2 scaffolds (40 kbp, 10 kbp), which could not be confidently placed. The chromosome contains the previously described acs1 operon, and two additional, undescribed acs operons (acs2, acs3), which differ from each other in gene content. For the chromosome, rings show from outside in: (1) read coverage, (2) coding sequences on forward and (3) reverse strands, (4) acs1, acs2 and acs3 operons, with their gene contents magnified, (5) GC percentage and (6) GC skew. Read coverage was similar across the genome (see outer ring), with an average RPKM value of 477. See Materials and Methods for details on sequencing and analysis. (B) Relatedness and amino acid percentage identity of AcsAB and AcsC proteins in the three acs operons. A phylogeny of AcsABC proteins suggests acs2 and acs3 to be more closely related to each other than the full acs1 operon. Furthermore, as acs2 and acs3 do not contain acsD, cmcax, ccpax nor bglxA, this suggests that they may have arisen in evolution as duplications of acs1, followed by gene loss. AcsAB proteins seem to be more conserved than AcsC, and share approximately 41–46% sequence identity, compared to 29–40% identity of AcsC. Amino acid sequences were aligned and percent identity calculated using MUSCLE54 and the tree built using the Neighbour-Joining method. All positions containing gaps were removed from analysis. Analyses were conducted using MEGA655.
