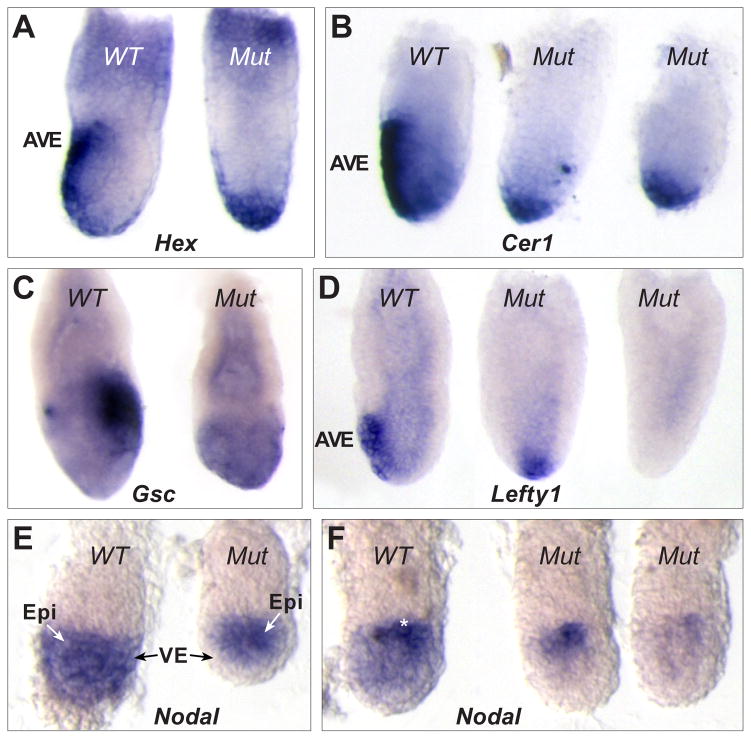
Fig. 4. AVE migration defects and reduced epiblast Nodal in Scheme 2 conditional mutants.
(A) Two E6.5 littermates after WMISH for Hex mRNA. The wild type embryo (left) shows Hex in AVE cells while the conditional mutant shows distal Hex expression. (B) WMISH for Cerl mRNA in three E6.5 littermates. The wild type embryo (left) shows Cerl expression in the AVE while the conditional mutants (center, right) show distal expression. (C) WMISH for Goosecoid (Gsc) mRNA in two E6.5 littermates. The wild type embryo (left) shows Gsc in primitive streak cells at the posterior. The conditional mutant (right) shows only background levels and is smaller and misshapen. (D) Three E6.5 littermates following WMISH for Lefty1 mRNA expression. The wild type embryo (left) shows Lefty1 in the AVE while the conditional mutants show either distal (center) or no expression (right). (E) WMISH for Nodal mRNA in two early E5.5 littermates. The wild type embryo (left) shows Nodal throughout the epiblast and VE. The conditional mutant (right) lacks Nodal expression in the VE. (F) WMISH for Nodal mRNA expression in three E5.5 littermates. The wild type embryo (left) shows reduced Nodal in the distal epiblast and VE and strong proximal expression (white asterisk). The conditional mutants (right, center) completely lack Nodal expression in the VE and distal epiblast and show reduced proximal expression compared to wild type.