Fig. 4.
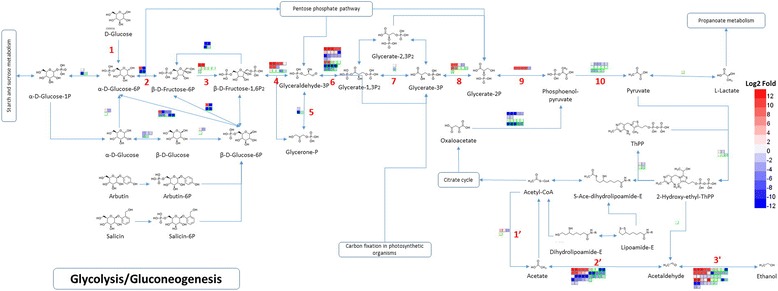
Differential expression of unigenes involved in glycolysis in E8 and E24 compared to C samples of Hevea brasiliensis. Glycolysis comprises 10 step reactions. In step 1, the enzyme hexokinase (HK) phosphorylates glucose by transferring a phosphate group from ATP to glucose forming glucose 6-phosphate. In step 2, glucose 6-phosphate is converted to its isomer fructose 6-phosphate by phosphoglucoisomerase (PGI). In step 3, the enzyme phosphofructokinase (PFK) catalyzes the conversion of fructose 6-phosphate into fructose 1,6-bisphosphate using another ATP to transfer a phosphate group to fructose 6-phosphate. In step 4, Fructose 1,6-bisphosphate aldolase (FBA) splits fructose 1,6-bisphosphate into dihydroxyacetone phosphate (DHAP, also glycerone phosphate) and glyceraldehyde 3-phosphate (G3P). In step 5, triose-phosphate isomerase (TPI or TIM) catalyzes the reversible interconversion of DHAP and G3P. In step 6, G3P is converted to 1,3-bisphosphoglycerate (1,3-BPG) by glyceraldehyde 3-phosphate dehydrogenase (GAPDH). In step 7, phosphoglycerate kinase (PGK) catalyzes the reversible transfer of the phosphate group from 1,3-BPG to ADP generating 3-phosphoglycerate (3-PG) and ATP. In step 8, the conversion of 3-PG to 2-phosphoglycerate (2-PG) is catalyzed by phosphoglycerate mutase (PGM). In step 9, enolase (or phosphopyruvate hydratase) catalyzes the dehydration of 2-PG to form phosphoenolpyruvate (PEP). In step 10, pyruvate kinase (PK) catalyzes the transfer of the phosphate group from PEP to ADP, yielding pyruvate and ATP. In an acetyl-CoA metabolic pathway, acetyl-CoA synthetase (ACS) (or acetate : CoA ligase) catalyzes the interconversion between acetyl-CoA and acetate (Step 1′), aldehyde dehydrogenase (ALDH) reversibly catalyze the conversion of acetate into acetaldehyde (Step 2′) and alcohol dehydrogenase (ADH) facilitates the interconversion between aldehydes and alcohols (Step 3′). Cells with gray border lines in the upper rows represent differentially expressed unigenes in E8 compared to C and cells with green border lines in the lower rows represent differentially expressed unigenes in E24 compared to C. Relative levels of expression are showed by a color gradient from low (blue) to high (red)
