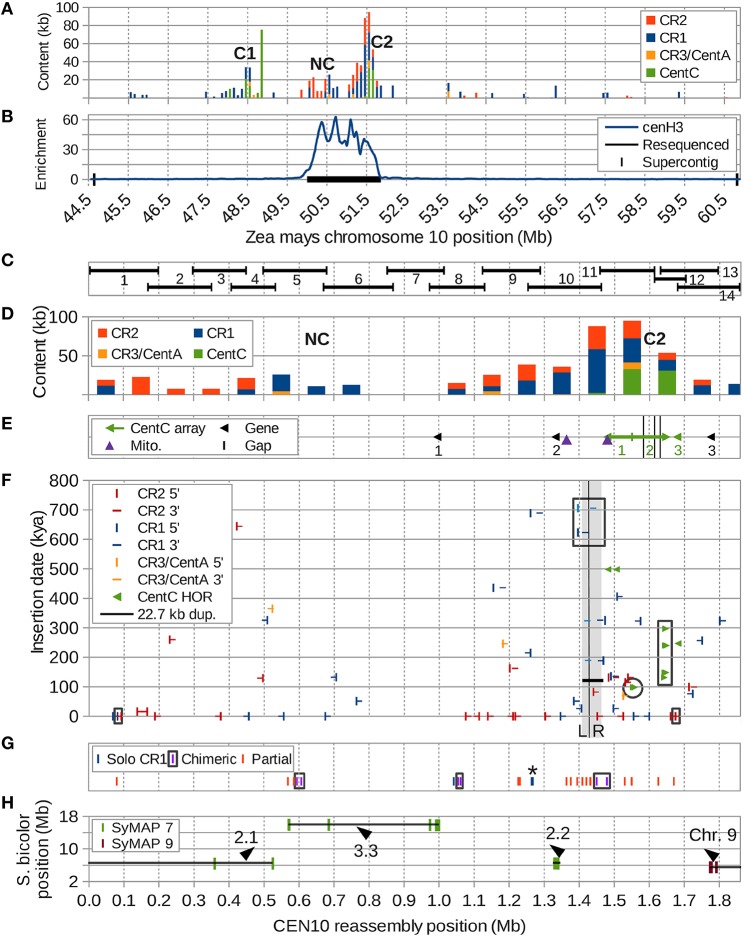
Figure 1.
Overview of CEN10 features. (A) Two centromeric repeat clusters containing CentC (C1 and C2) in the CEN10-containing supercontig were split by an ancestral hemicentric inversion and are now separated by a 2.6 Mb region including a CR cluster with no CentC (NC). CR3/CentA means CR3 and CentA (nonautonomous CR3). (B) Enrichment of anti-cenH3 ChIP-seq reveals NC and C2 are in the active centromere. The resequenced region is indicated by the black horizontal bar. (C) The regions of the resequenced CEN10 spanned by each BAC are indicated. (D) Centromeric repeat content per 100 kb window. (E) Positions of the three CentC arrays (#1 and #2 are interrupted by retrotransposons), three transcribed genes (probed by FISH), two mitochondrial insertions and three remaining sequence gaps (black vertical bars within CentC array #2). Arrows indicate the orientation of the CentC arrays; the junction of two adjoining inverted arrays is marked by a vertical bar. Arrowheads indicate direction of transcription for the three genes. (F) Estimated dates of CR element insertion, duplication of the 22.7 kb region (horizontal black line, the duplication is marked by adjacent gray segments labeled “L” and “R” and separated by a vertical black line) and seven higher-order repeats (HORs) of CentC (paired, mostly overlapping, green arrows) are indicated on the y axis. Three CRs with internal recombinations (including a CR partially in the 22.7 kb duplication) are boxed (black). The youngest of the dated CentC HORs (Figure 5) is circled, HORs from Figure 6 are boxed. (G) Positions of recombinant CR sequences, including three solo CR1 LTRs (* = two solos separated by only 2.3 kb), three chimeric elements (complete but with mismatched TSDs, LTR pairs are boxed) and 14 partial CR elements. For clarity, only the two closest ends of a chimeric element located in the 22.7 kb duplication are shown. A partial CR1 that may be artificially truncated by BAC (#12) vector is shown at position 1.63 Mb. (H) Synteny markers shared with sorghum chromosomes 7 (SyMAP 7) and 9 (SyMAP 9) are indicated by their sorghum and maize coordinates with orientation of blocks (lines) indicated by arrowheads. The most downstream marker cluster of the second synteny block (at 1 Mb) is inverted relative to sorghum, but co-linear with Brachypodium and rice. The 1.85 Mb CEN10 is composed of four different syntenic segments from two different sorghum chromosomes indicated here with labels as described in Figure 7 and Supplemental Figure S10.