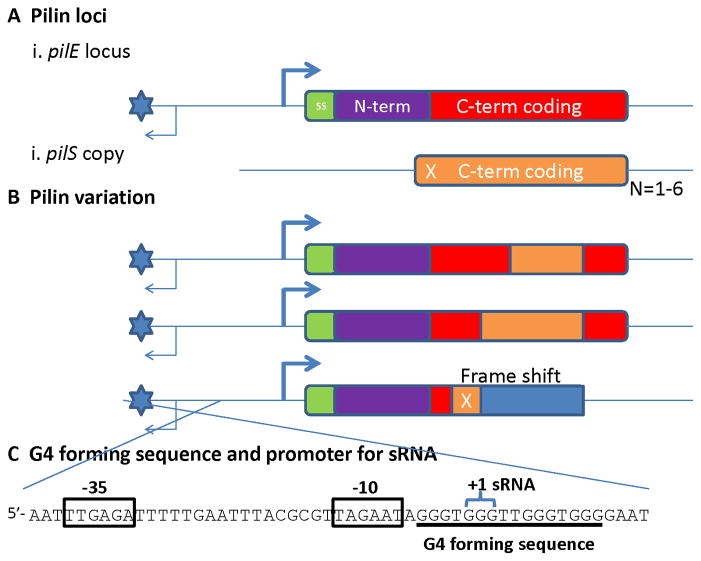
Figure 11. Neisserial pilin antigenic variation.
A) The pilin loci. (i) The pilin expression locus (pilE) has a promoter sequence to initiate transcription, a 7 amino acid signal sequence (SS – green); an absolutely-conserved, N-terminal constant coding sequence (N-term, purple) and a variable carboxy-terminal coding sequence (C-term, red). The G4 forming sequence is represented by the blue star and the sRNA transcript by the thin arrow. (ii) Shown is a variant pilS gene copy that has variant pilin C-terminal coding sequences (C-term, orange), but cannot express a protein product. This pilS copy has a frame shift in the 5′-coding sequences (X). Each pilS locus can have one to six repeated pilS copies (N=1–6). B) Pilin variation. Nonreciprocal transfer of variant pilS copy sequences into the pilE locus produces pilin variants. The pilS sequences that are transferred can be from any part of the pilS copy that overlaps with the pilE sequence as long as there are regions of microhomology at the ends of the transferred sequence (not shown). Some (but not all) pilS copies encode a frame shift in their 5′ potential coding sequences that can result in an altered reading frame (blue), a premature stop codon and a nonpiliated variant. C) The G4 forming sequence and promoter for the cis-acting sRNA. The sequence shown is on the bottom strand of the cartoons in parts A and B. The sRNA promoter is indicated by the boxed −10 and −35 sequences. The sRNA required for pilin antigenic variation initiates (+1 sRNA) within the second set of Gs in the G4 forming sequence.