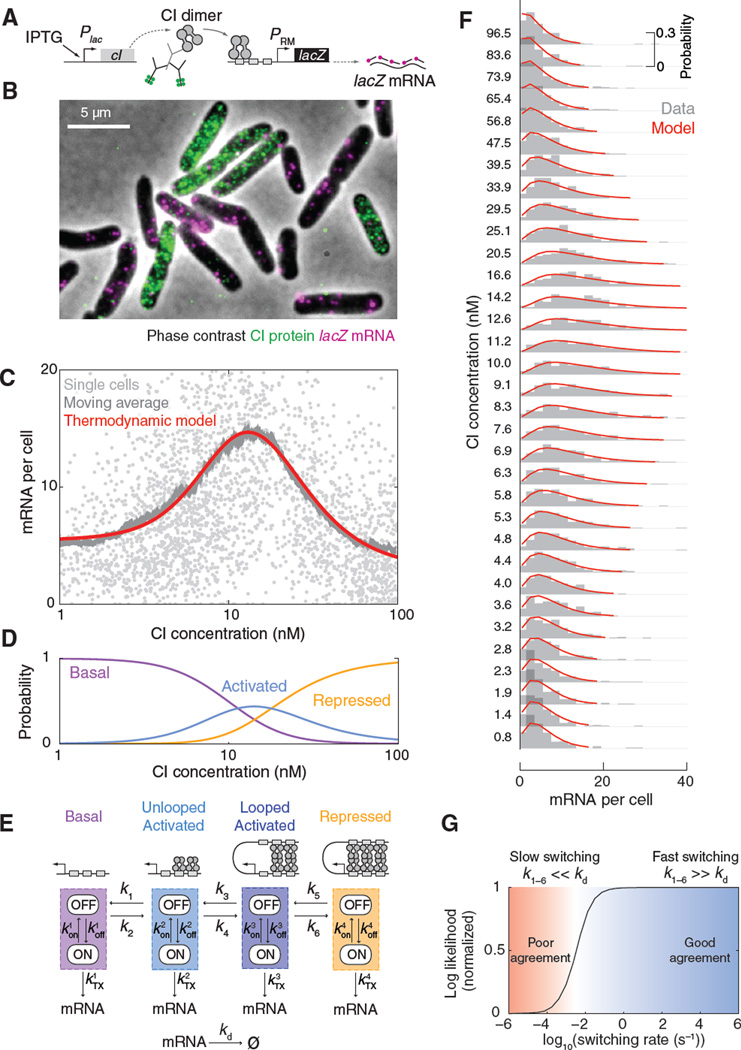
Fig. 4. PRM regulation at the single-cell level.
(A) The reporter system used for measuring PRM activity. (B) Cells labeled for CI protein (immunofluorescence, green) and PRM-lacZ mRNA (smFISH, magenta). (C) The PRM(CI) gene regulation function. The single-cell data (light gray, N = 2941 cells) was filtered using a moving average (dark gray, 200 cells/bin). The averaged curve was well fitted by the thermodynamic model (red). (D) The calculated probabilities of different promoter activity states as a function of CI concentration: basal (purple), activated (looped and unlooped, blue) and repressed (orange). (E) A stochastic model for PRM kinetics. Each promoter activity state was modeled using stochastic ON/OFF transcription kinetics. The promoter stochastically transitions between activity states in a CI-dependent manner. (F) The stochastic model successfully described the PRM(CI) single-cell data. The experimental data (panel C) was binned into 100-cell histograms (gray) and fitted to the model (red) using maximum likelihood estimation. (G) Consistency of measured mRNA statistics with rapid switching between promoter states. Solving the stochastic model for different switching rates, and fitting the model results to the measured mRNA statistics, resulted in a good fit for fast switching (right, blue); slow switching yielded a poor fit (left, red).